GOAT: Gene-level biomarker discovery from multi-Omics data using graph ATtention neural network for eosinophilic asthma subtype
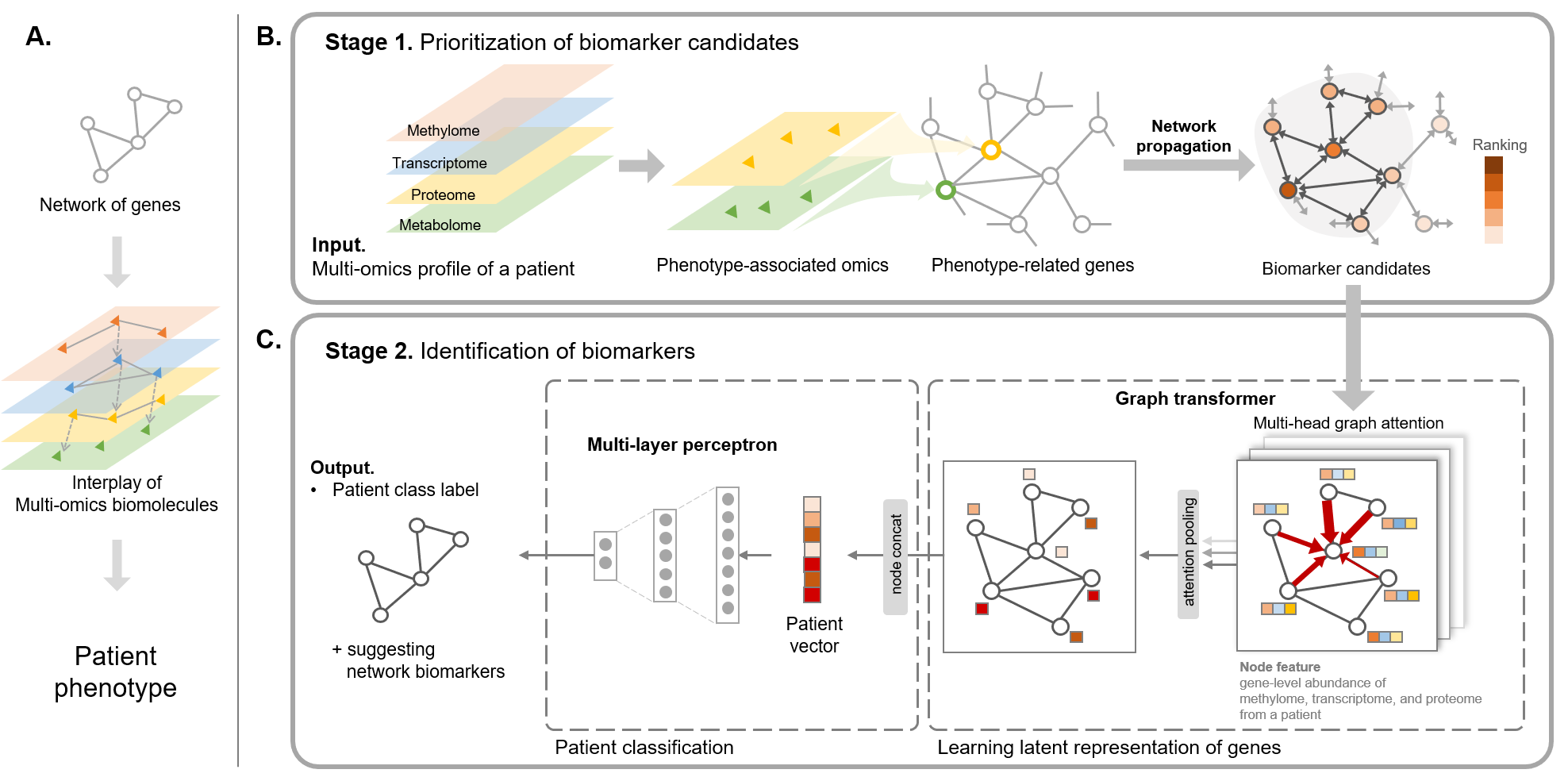
We propose a novel deep graph attention model for biomarker discovery for the asthma subtype by incorporating complex interactions between biomolecules and capturing key biomarker candidates using the attention mechanism.
You can build a docker image from Dockerfile.
# Pull base image from docker hub
docker pull dabinjeong/cuda:10.1-cudnn7-devel-ubuntu18.04
# Build docker image
docker build --tag biomarker:0.1.1 .
You can also download the docker image from Docker hub (https://hub.docker.com/repository/docker/dabinjeong/biomarker/general).
docker pull dabinjeong/biomarker:0.1.1
conda create -n biomarker python=3.9
conda activate biomarker
conda install -c bioconda nextflow=21.04.0
nextflow run biomarker_discovery.nf -c pipeline.config -with-docker biomarker:0.1.1
For comparative analysis, please refer to the following repository, comparative_analysis_multi-omics_biomarker.