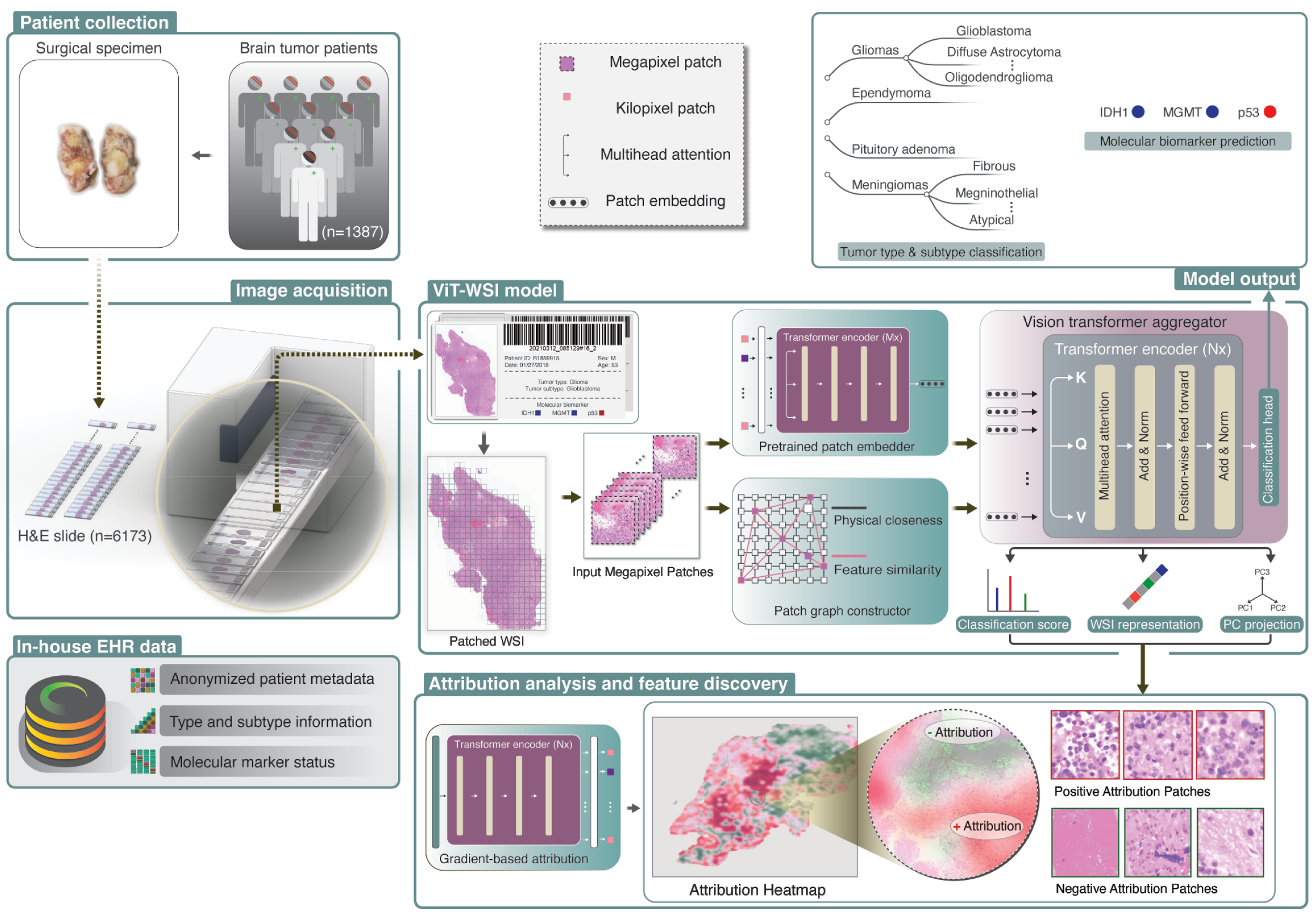
ViT-WSI: Vision Transformer-based Weakly-Supervised Histopathological Image Analysis of Primary Brain Tumors
This repository contains the code of ViT-WSI from
Li,Zhongxiao, et al. "Vision Transformer-based Weakly-Supervised Histopathological Image Analysis of Primary Brain Tumors".
The code is tested with the following dependencies:
- Python related dependencies
- python 3.9
- cython 0.29.24
- Pytorch related dependencies
- pytorch 1.8.2
- torchvision 0.9.2
- tensorboardX 2.4
- einops 0.3.2
- timm 0.4.12
- numpy 1.19.5
- scipy 1.7.1
- pandas 1.3.2
- pyTables 3.5.2
- h5py 3.1
Modify the information of conda environment in shell_scripts/setup_environment.sh to let it be used in meta-main.sh.
An example glioma dataset containing 147 glioma slides from TCGA is available at dataset_files/TCGA-glioma-subtyping/info/selected_slides-info.csv.
Please download the corresponding slides (in svs format) from The Cancer Genome Atlas (TCGA), and save them to dataset_files/TCGA-glioma-subtyping/WSI.
In the following steps, the intermediate results will be saved to the subdirectories of dataset_files/TCGA-glioma-subtyping.
To extract 1024×1024 patches from the slides:
bash meta-main.sh create_patches
The results will be saved to dataset_files/TCGA-glioma-subtyping/patches.
To extract features using the feature extractor ViT-L-16:
bash meta-main.sh extract_feature
The results will be saved to dataset_files/TCGA-glioma-subtyping/features.
Compile the Cython extention:
python setup.py build_ext --inplace
Build graph that will be automatically saved to pickle files:
bash meta-main.sh build_slide_graph
The results will be saved to dataset_files/TCGA-glioma-subtyping/slide_graph.
bash meta-main.sh create_splits
The dataset will be randomly split into 10 folds. In each fold, #train:#test=8:2.
The results will be saved to dataset_files/TCGA-glioma-subtyping/splits.
To run the ViT-WSI aggregater training without slide graph, use TCGA-glioma-subtyping-vit_aggr as the experimental code. This uses config_files/TCGA-glioma-subtyping-vit_aggr.yaml as the configuration file.
bash meta-main.sh train_eval TCGA-glioma-subtyping-vit_aggr
To run the ViT-WSI aggregater training with slide graph, use TCGA-glioma-subtyping-graph_vit_aggr as the experimental code. This will use config_files/TCGA-glioma-subtyping-graph_vit_aggr.yaml as the configuration file.
bash meta-main.sh train_eval TCGA-glioma-subtyping-graph_vit_aggr
This aggregates the performance statistics across the folds. Depending the experiment that has been performed, either use
bash meta-main.sh eval_aggr TCGA-glioma-subtyping-vit_aggr
or
bash meta-main.sh eval_aggr TCGA-glioma-subtyping-graph_vit_aggr
The performance statistics will be saved to dataset_files/TCGA-glioma-subtyping/train/TCGA-glioma-subtyping-graph_vit_aggr/fold_aggr.
The preprocessing part (patch creation, feature extraction, and dataset splitting) was developed based on the code of CLAM. We thank the original authors for making their code publicly accessible to the community.