DREDDA: Drug Repositioning through Expression Data Domain Adaptation
Overview
This repository contains the implementation of DREDDA from
Zhongxiao Li, Antonella Napolitano, Monica Fedele, Xin Gao*, Francesco Napolitano*
"AI identifies potent inducers of breast cancer stem cell differentiation based on adversarial learning from gene expression data" bioRxiv, 2023
If you use our work in your research, please cite our paper, which is now available on bioRxiv
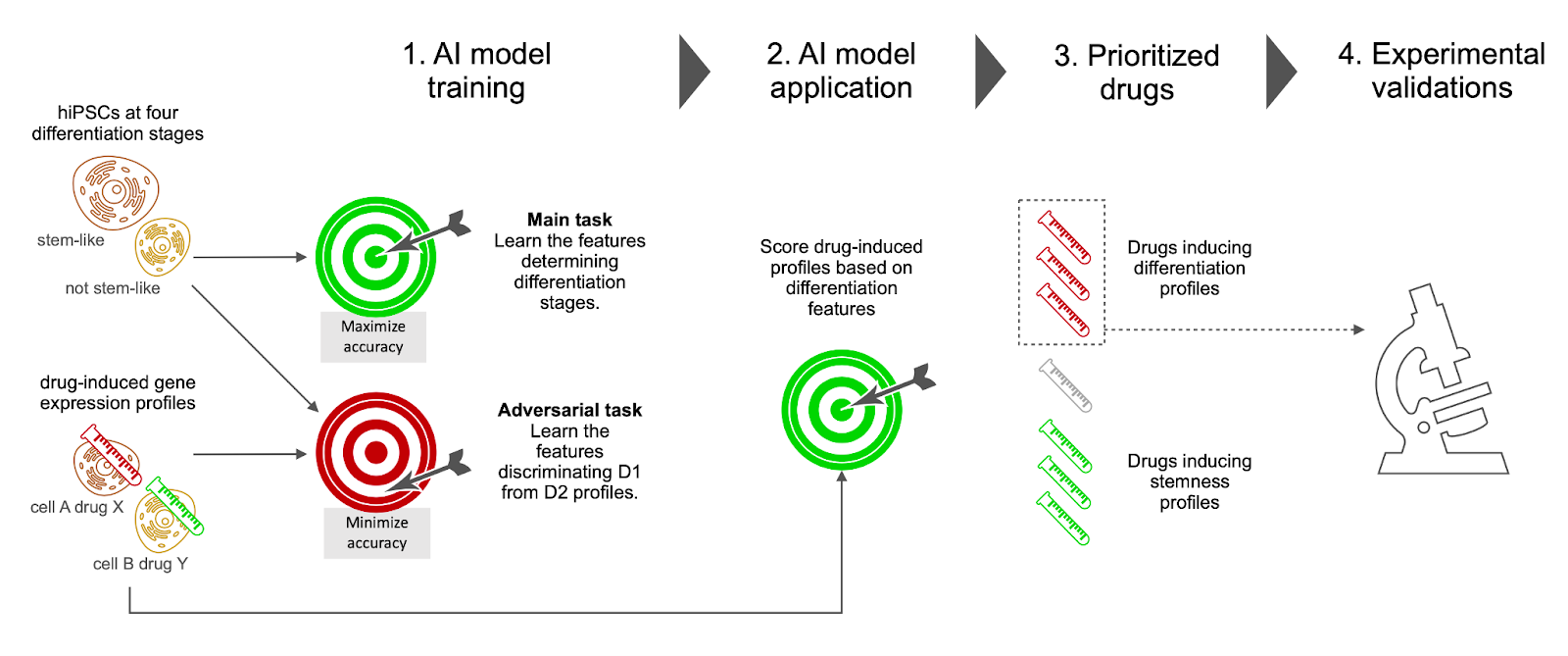
Fig. 1 Overview of the study. Single-cell gene expression profiles of hIPSCs at various differentiation stages and drug-induced gene expression profiles were fed to an adversarial learning model, which simultaneously learned differentiation features to be used in subsequent predictions (main task) and dataset specific features to be avoided (adversarial task). The trained model was then used to score all the drug-induced profiles. A selection of 6 drugs among the top scoring ones was experimentally validated.
The architecture of DREDDA
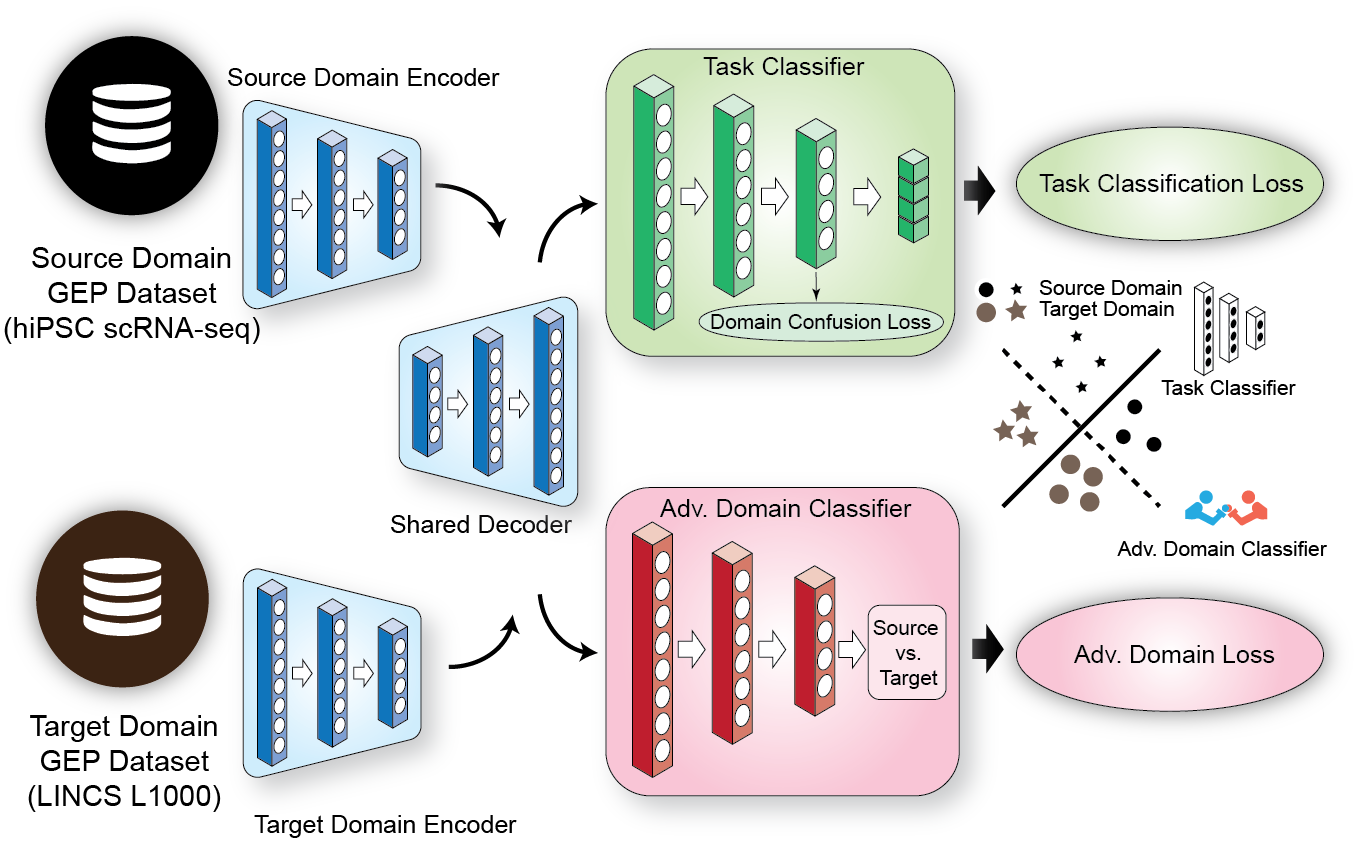
Fig. 2 The DREDDA model architecture includes one encoder for each dataset and a shared decoder; the resulting profiles from the source domain are sent to the main task classifier (positively weighted in the overall loss function), while both source and target domain profiles are sent to the adversarial classifier (negatively weighted).
Prerequisites
The code is tested with the following dependencies:
- Python 3.7.10
- Pytorch 1.6.0
- Torchvision 0.7.0
- Numpy 1.20.2
- Scipy 1.6.2
- Pandas 1.2.4
- PyTables 3.6.1
- h5py 2.10.0
Reproducing the LINCS drug list
To run DREDDA with our pretrained checkpoint, simply use
python main-DREDDA.py test
Relavant files will be downloaded to download/. The results will (by default) be saved to train_dir/default_config/test.
Training from scratch
To train DREDDA with the default configuration, simply use
python main-DREDDA.py train
Relavant files will be downloaded to download/. The resulting checkpoints will be saved to train_dir/default_config/
The following example shows how to change the default behavior
python main-DREDDA.py train \
--source_dataset_name CRISPRi \
--target_dataset_name LINCS \
--out_dir train_dir/default_config \
--n_epochs 200 \
--dual_training_epoch 150 \
--domain_adv_coeff 0.1 \
--ddc_coeff 0.1 \
--seed 41
To test a specific checkpoint, please use
python main-DREDDA.py test --ckpt_fp <path_to_checkpoint>