This repository contains the manuscript mentioned in the title, and associated code and data sets. Should you need help running our code, please contact us.
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2175-5
Bardozzo, Francesco, Pietro Lió, and Roberto Tagliaferri. "A study on multi-omic oscillations in Escherichia coli metabolic networks." BMC Bioinformatics 19.7 (2018): 194.
Background: The two important challenges in the analysis of molecular biology information are the following: (a) data (multi-omic information) integration; (b) detecting patterns across large scale molecular networks. The two challenges are actually coupled as the integration of omic information may provide better means to detect multi-omic patterns that could reveal multi-scale or emerging properties at the phenotype levels.
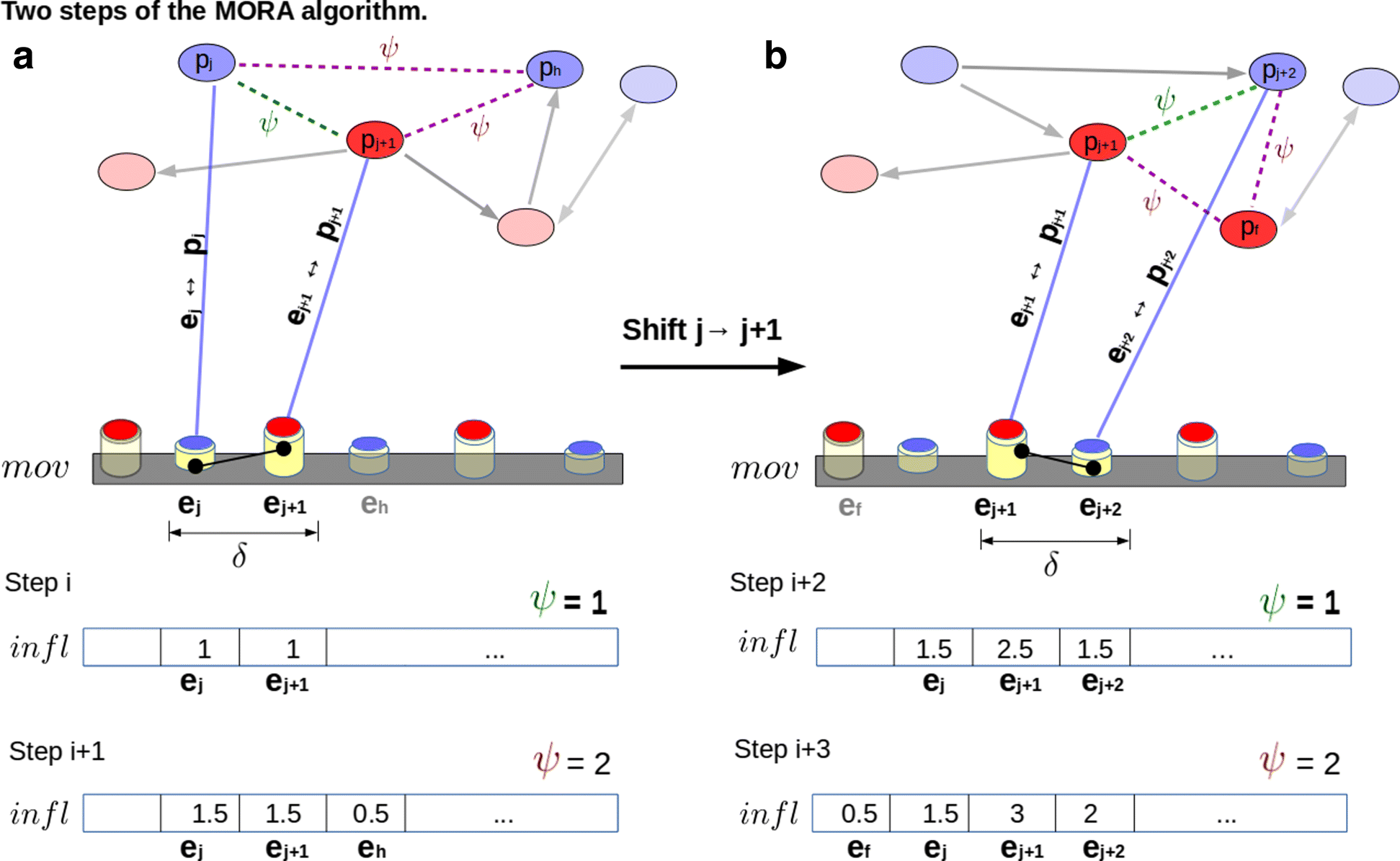
Results: Here we address the problem of integrating various types of molecular information (a large collection of gene expression data, sequence data, codon usage and protein abundances) to analyse the Escherichia Coli metabolic response to treatments at the entire network level.Our algorithm, MORA (Multi-omic relational adjacency) is able to detect patterns which may represent metabolic network motifs at pathway and supra pathway levels which could hint at some functional role. We provide description and insights on the algorithm by testing it on a large database of responses to antibiotics. Along with the MORA algorithm, a novel model for the analysis of alternating multi-omics has been proposed. Interestingly, the resulting analysis suggests that some motifs reveal recurring alternating or position variation patterns on multi-omics metabolic networks. Our framework, implemented in R, provides effective and friendly means to design intervention scenarios on real data. By analysing how multi-omics data build up multi-scale phenotypes, the software allows to compare and test metabolic models, design new pathways or redesign existing metabolic pathways and validate in silico metabolic models using nearby species.
Conclusions: The integration of multi-omic data reveals that bacterial multi-omic metabolic networks contain position dependent and recurring patterns which could provide clues of long range correlations in the bacterial genome. Source Code Source for the MORA is in the main repository, and source used to generate multi-omic pattens. The E.coli whole metabolic network is integrated from KEGG and EcoCyc.
Here, for the impatient, is an implementation of MORA in R
MORA.comp <- function(pattern, delta, psi, g)
{
couple <- length(pattern) - (delta - 2)
dir.edges <- "all"
count <- c(rep(0, length(pattern)))
names(count) <- as.character((pattern))
count <- c(count, count[1:max(psi) - 1])
pattern <- c(pattern, pattern[1:max(psi) - 1])
for (y in 1:length(psi))
{
for (i in 1:couple)
{
p.from <- pattern[i]
p.to <- pattern[i + (delta - 1)]
patty <- NULL
if (distances(g, v = p.from, to = p.to, mode = dir.edges)[[1]] !=
Inf)
{
tryCatch(patty <- get.shortest.paths(g, p.from, p.to, mode = dir.edges,
weights = NULL, output = c("vpath", "epath", "both")),
warning = function(w)
{
print(paste("warning... "))
}, error = function(e)
{
print(paste("no path. "))
})
}
if (length(patty) != 0)
{
for (k in 1:length(patty$vpath))
{
if (length(V(g)[patty$vpath[[k]]]$name) == psi[[y]])
{
pr <- paste("Pattern elements delta = ", delta - 1,
". Vertex psi neighbouring = ", psi[[y]] - 1, "edges.",
sep = " ")
pr <- paste(pr, " Shortest path's end nodes: ", names(count)[i],
" and ", names(count)[i + (delta-1)], sep=" ")
namenodes <- V(g)[patty$vpath[[k]]]$name
for (z in 1:length(namenodes))
{
zx <- grep(paste("\\b", namenodes[z], "\\b", sep = ""),
names(count))
print(paste("", namenodes))
count[c(zx)] <- (count[c(zx)] + (1/(psi[[y]] - 1)))
print(paste("", count[c(zx)]))
}
}
}
}
}
}
count <- count[!duplicated(names(count))]
return(count)
}
Licence BMC Bioinformatics, This work is supported by the Computer Laboratory (University of Cambridge, UK), NeuRoNe Lab (University of Salerno - IT)
** .BIB ** @Article{Bardozzo2018, author="Bardozzo, Francesco and Li{'o}, Pietro and Tagliaferri, Roberto", title="A study on multi-omic oscillations in Escherichia coli metabolic networks", journal="BMC Bioinformatics", year="2018", month="Jul", day="09", volume="19", number="7", pages="194", issn="1471-2105", doi="10.1186/s12859-018-2175-5", url="https://doi.org/10.1186/s12859-018-2175-5" }