GBSON
File format definition used by the GeSeq annotation software to represent GenBank data as JSON.
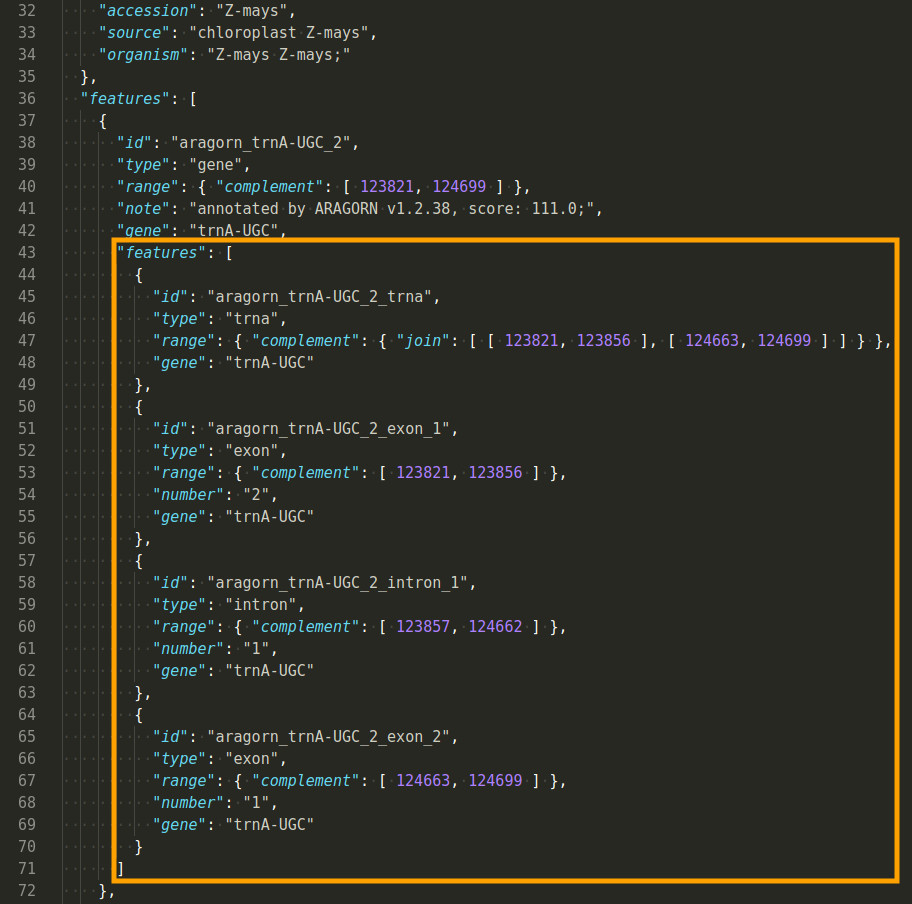
Example JSON output
An example output can be found here: GBSON-Example.json.
Usage
To use GBSON in your application you can use any standard JSON parser. If you are programming in JavaScript, just do
const data = JSON.parse(gbson);In TypeScript you should add the type declaration from this repository to get full IDE support
import { GBSON } from "./GBSON";
const data:GBSON = JSON.parse(gbson);And access the data like this
const gene = data.features[0].gene;Range definitions
GenBank
complement(join(125294..125832,126877..127429))
GBSON
{ "complement" : { "joined" : [ [ 125294, 125832 ], [ 126877, 127429 ] ] } }Nested Features
An advantage of using JSON over GenBank or GFF is its intrinsic capability to express nested structures: