IIB-MIL: Integrated instance-level and bag-level multiple instances learning with label disambiguation for pathological image analysis
Code for paper titled "IIB-MIL: Integrated instance-level and bag-level multiple instances learning with label disambiguation for pathological image analysis" submitted to MICCAI2023. The basic method and applications are introduced as follows:
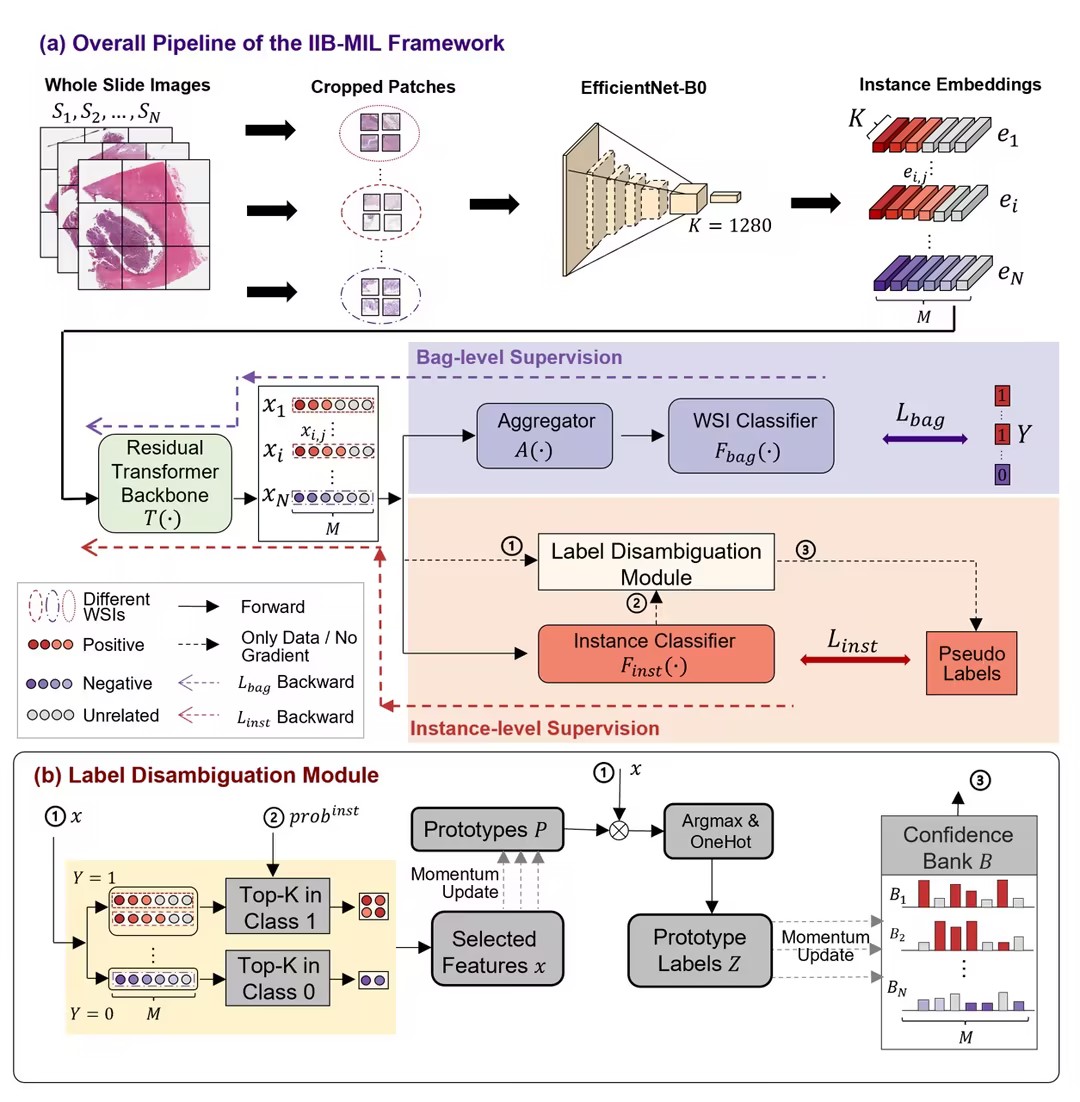
IIB-MIL first transforms input huge-size WSI to a set of patch embeddings to simplify the following learning task using a pre-trained encoder, i.e. EfficientNet-B0. Then a specially-designed residual transformer backbone works to calibrate the obtained patch embeddings and encode the context information and correlation of patches. After that, IIB-MIL utilizes both a transformer-based bag-level and a label-disambiguation-based instance-level supervision to cooperatively optimize the model, where the bag-level loss is calculated referring to the WSI labels, while the instance loss is calculated referring to pseudo patch labels calibrated by the Label-Disambiguation module. Since bag-level supervision channel is trained to globally summarise information of all patches for prediction, the bag-level outputs are used as the final predictions during the test stage.
albumentations==1.1.0
efficientnet_pytorch==0.7.1
lifelines==0.26.4
numpy==1.21.6
opencv_contrib_python==4.8.0.74
opencv_python==4.4.0.46
opencv_python_headless==4.5.4.60
openslide_python==1.2.0
pandas==1.3.5
Pillow==8.4.0
pretrainedmodels==0.7.4
rich==10.16.1
scikit_image==0.19.1
scikit_learn==1.0.1
Shapely==1.7.1
torch==1.7.1
torchvision==0.8.2
tqdm==4.62.3
yacs==0.1.8
The preprocessed test data of TCGA-NSCLC and TCGA-RCC Dataset is available on Zenodo at: https://zenodo.org/record/8169144.
This code uses the centralized configs. Before using this code, a config file needs to be edited to assign necessary parameters. The config files are provided for different Datasets:
configs/LUAD-GM.yaml
configs/TCGA-NSCLC.yaml
configs/TCGA-RCC.yaml
1、First, in order to split the WSI into patches, execute the following script.
python ./data_prepare/WSI_cropping.py \
--dataset path_to_data_folder \
--output path_to_output_folder \
--scale 20 --patch_size 1120 --num_threads 16
2、Then, extract features from each patch. a pre-trained feature extractor can be utilized here (e.g. EfficientNet-B0 trained on the ImageNet).
python ./data_prepare/extract_feature.py --cfg cconfigs/LUAD-GM.yaml
# or
python ./data_prepare/extract_feature.py --cfg cconfigs/TCGA-NSCLC.yaml
# or
python ./data_prepare/extract_feature.py --cfg cconfigs/TCGA-RCC.yaml
3、Next, combine features of one WSI.
python ./data_prepare/merge_patch_feat.py --cfg configs/LUAD-GM.yaml
# or
python ./data_prepare/merge_patch_feat.py --cfg configs/TCGA-NSCLC.yaml
# or
python ./data_prepare/merge_patch_feat.py --cfg configs/TCGA-RCC.yaml
4、Finally, we can use the model with preprocessed data
python ./main/test.py --cfg configs/LUAD-GM.yaml --input_data_file data/LUAD-GM.txt
# or
python ./main/test.py --cfg configs/TCGA-NSCLC.yaml --input_data_file data/TCGA-NSCLC.txt
# or
python ./main/test.py --cfg configs/TCGA-RCC.yaml --input_data_file data/TCGA-RCC.txt
This repo partially uses code from Deformable DETR, T2T-ViT and [iRPE] (https://github.com/microsoft/Cream.git).