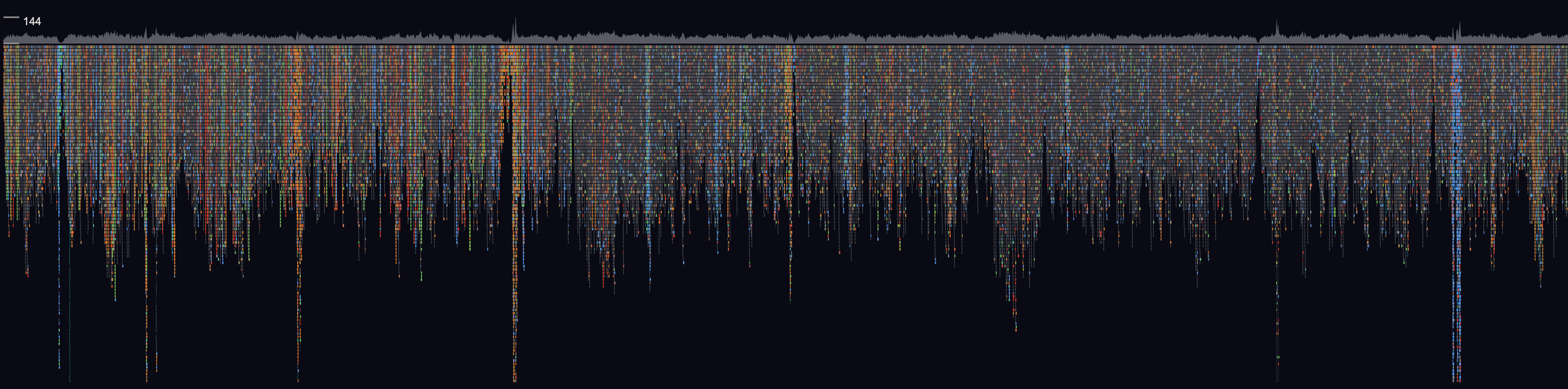
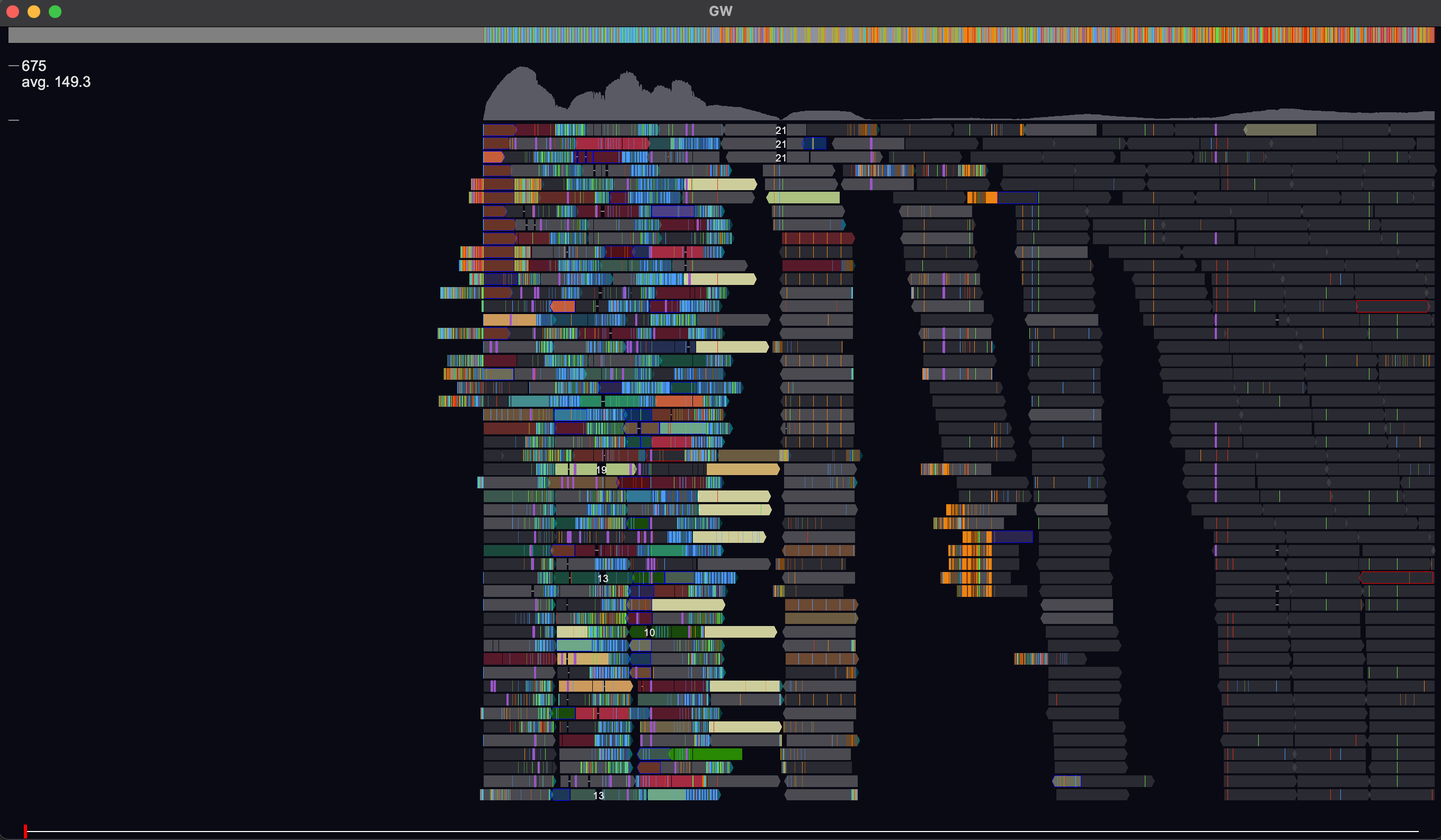
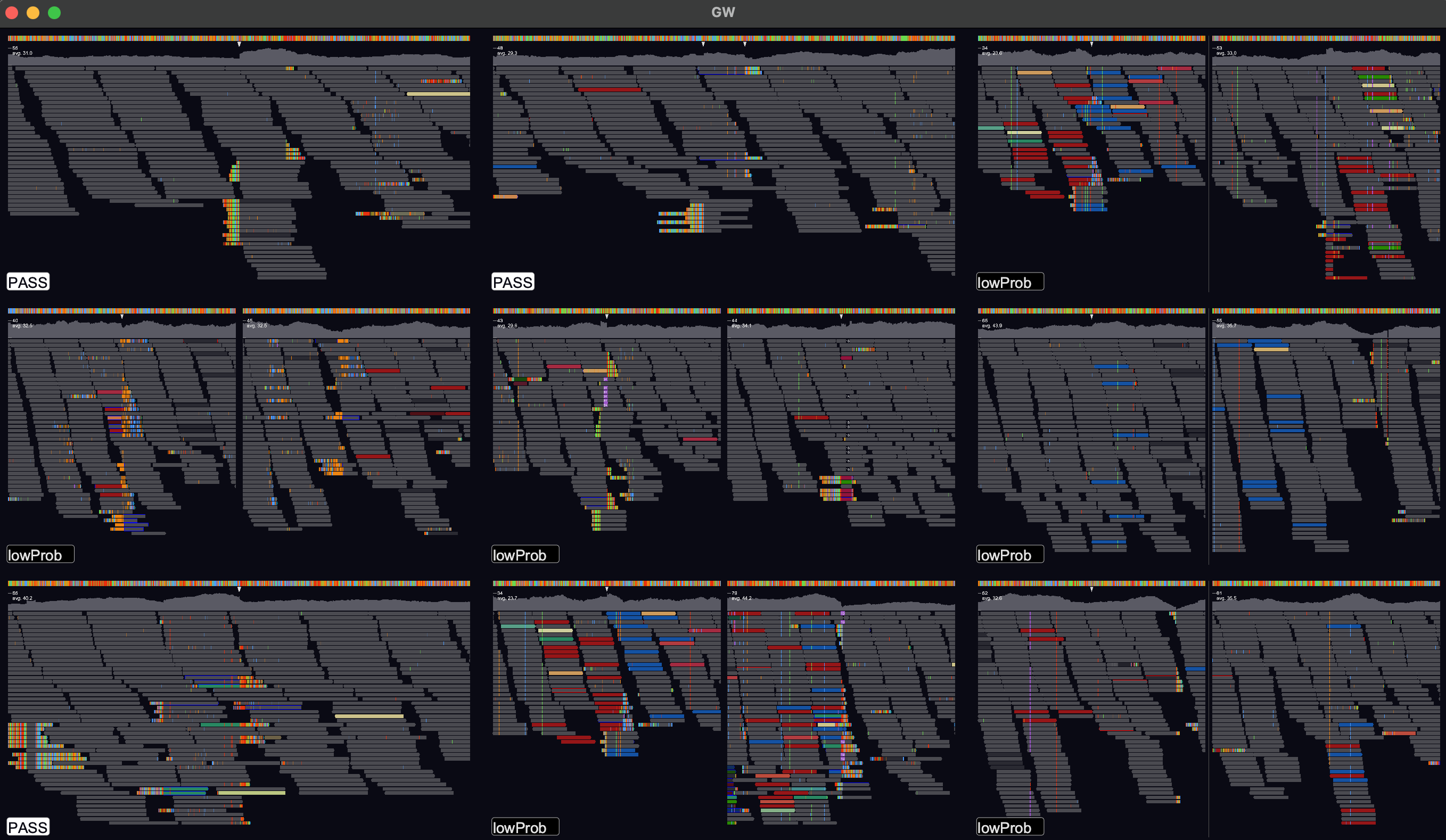
GW is a fast browser for genomic sequencing data (.bam/.cram format) used directly from the terminal. GW also allows you to view and annotate variants from vcf/bcf files.
Check out the documentation here.
For best performance, download one of the app packages from the Releases page. See the install section of the documentation for more details.
Using a package manager:
conda install -c bioconda -c conda-forge gw
brew install kcleal/homebrew-gw/gw
To build from source, using conda to fetch htslib + glfw3 dependencies:
conda create -y -n gw_env -c conda-forge glfw htslib conda activate gw_env git clone https://github.com/kcleal/gw.git cd gw && make prep CONDA_PREFIX=$(conda info --base) LDLIBS+="-lcrypto -lssl" make -j4
Command line:
# Start gw (drag and drop bams into window) gw hg38 # View start of chr1 gw hg38 -b your.bam -r chr1 # Two regions, side-by-side gw hg38 -b your.bam -r chr1:1-20000 -r chr2:50000-60000 # Multiple bams gw hg38 -b '*.bam' -r chr1 gw hg38 -b b1.bam -b b2.bam -r chr1 # Add a track BED/VCF/BCF/LABEL gw hg38 -b your.bam -r chr1 --track a.bed # png image to stdout gw hg38 -b your.bam -r chr1:1-20000 -n > out.png # Save pdf gw hg38 -b your.bam -r chr1:1-20000 -n --fmt pdf -f out.pdf # plot every chromosome in parallel gw t2t -t 24 -b your.bam -n --outdir chrom_plots # View VCF/BCF gw hg38 -b your.bam -v var.vcf # View VCF/BCF from stdin gw hg38 -b your.bam -v - # View some png images gw -i "images/*.png" # Save some annotations gw hg38 -b your.bam -v var.vcf --labels Yes,No --out-labels labels.tsv
Here are a few GW commands (others are available). Access command box with : or /:
help # help menu config # open config file for editing chr1:1-20000 # Navigate to region add chr2:1-50000 # Append new region rm 1 # Region at column index 1 removed rm bam1 # Bam file at row index 1 removed mate # Move view to mate of read mate add # mate added in new view line # Toggle vertical line ylim 100 # View depth increased to 100 find QNAME # Highlight all reads with qname==QNAME filter mapq >= 10 # Filer reads for mapq >= 10 count # Counts of all reads for each view point snapshot # Save screenshot to .png man COMMAND # manual for command
To view a genomic region e.g. chr1:1-20000, supply an indexed reference genome and an alignment file (using -b option):
gw hg38 -b your.bam -r chr1:1-20000
A variant file in .vcf/.bcf format can be opened in a GW window by either dragging-and-dropping or via the -v option:
gw hg38.fa -b your.bam -v variants.vcf
See test directory.
If you find bugs, or have feature requests please open an issue, or drop me an email clealk@cardiff.ac.uk. GW is under active development, and we would welcome any contributions!