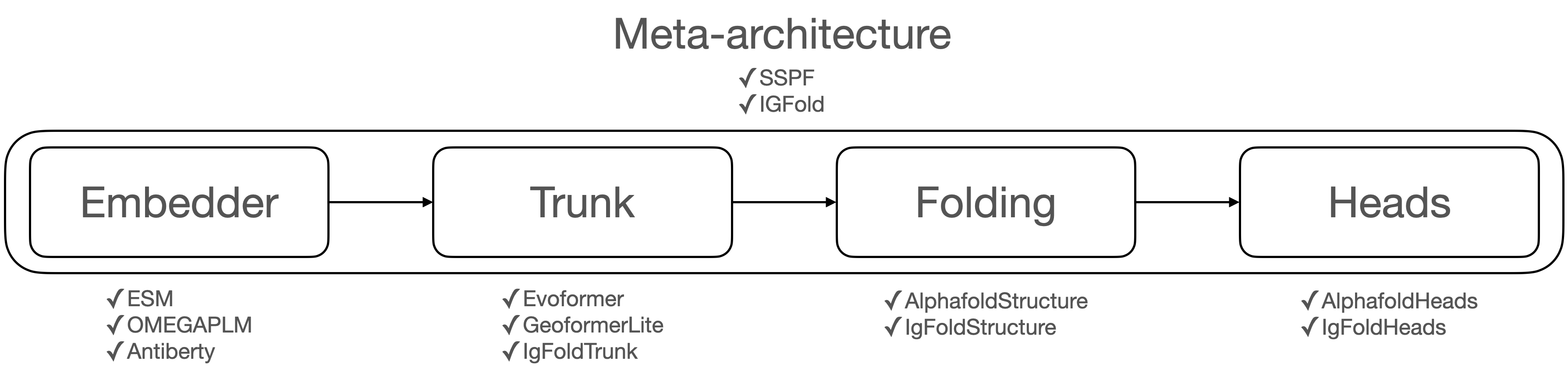
Solvent is a library that provides protein folding algorithms. It supports single sequence based protein folding including ESMFold, OmegaFold, and IgFold. Researchers can train and evaluate each model with same conditions and design new model variant by combining modules.
See data preparation
Training ESMFold on single GPU
# initial training
python train_net.py \
--config-file configs/esm35_evo1_initial_pdbonly.yaml \
--num-gpus 1 SOLVER.SEQ_PER_BATCH 2 \
OUTPUT_DIR output/esm35_evo1/initial_pdbonly
# finetuning from initially trained model
python train_net.py \
--config-file configs/esm35_evo1_finetune_pdbonly.yaml \
--num-gpus 1 SOLVER.SEQ_PER_BATCH 2 \
OUTPUT_DIR output/esm35_evo1/finetune_pdbonly \
MODEL.WEIGHTS output/esm35_evo1/initial_pdbonly/model_final.pth
Training models using DDP
# e.g. 16 batch with 2 machines(8GPU)
# (machine 0)
python train_net.py \
--config-file configs/esm35_evo1_initial_pdbonly.yaml \
--num-gpus 8 --num-machines 2 --machine-rank 0 --dist-url <URL> \
SOLVER.SEQ_PER_BATCH 16 \
OUTPUT_DIR output/esm35_evo1/initial_pdbonly
# (machine 1)
python train_net.py \
--config-file configs/esm35_evo1_initial_pdbonly.yaml \
--num-gpus 8 --num-machines 2 --machine-rank 1 --dist-url <URL> \
SOLVER.SEQ_PER_BATCH 16 \
OUTPUT_DIR output/esm35_evo1/initial_pdbonly
Evaluation on trained model
python train_net.py \
--eval-only \
--config-file output/esm35_evo1/finetune_pdbonly/config.yaml \
--num-gpus 1 \
MODEL.WEIGHTS output/esm35_evo1/finetune_pdbonly/model_final.pth
Inference from fasta
python demo/demo.py \
--config-file output/esm35_evo1/finetune_pdbonly/config.yaml \
--input datasets/cameo/fasta_dir/* \
--output output/esm35_evo1/finetune_pdbonly/results \
--opt \
SOLVER.SEQ_PER_BATCH 1 \
MODEL.WEIGHTS output/esm35_evo1/finetune_pdbonly/model_final.pth
This repository is heavily depend on the project listed below.
To make Solvent working as framework, we refer the pipeline of Detectron2. We represent individual method using the implementation of AlphaFold2, OpenFold, IgFold, and OmegaFold.
We acknowledge the contributions of the Language Model Engineering Team at Kakao Brain, who have optimized Solvent. These optimizations make Solvent efficient in training speed and memory, so researchers can easily tap larger models. Their support has been essential in achieving the outcomes presented in this work.
The description of Solvent is in the technical report below.
@misc{lee2023solvent,
title={Solvent: A Framework for Protein Folding},
author={Jaemyung Lee and Kyeongtak Han and Jaehoon Kim and Hasun Yu and Youhan Lee},
year={2023},
eprint={2307.04603},
archivePrefix={arXiv},
primaryClass={q-bio.BM}
}