This repository contains supplementary data and code used for the manuscript "Understanding drivers of phylogenetic clustering and terminal branch lengths distribution in epidemics of Mycobacterium tuberculosis" (Menardo 2022, eLife2022;11:e76780 DOI: https://doi.org/10.7554/eLife.76780).
- sim_results: a folder containing results for all simulations performed in the study (clustering rates and terminal branch lengths)
- MTB_cluster_sim.py: the python wrapper fro the pipeline. The pipeline was tested on Ubuntu and CentOS.
- collect_res.R: R script to join outputs (after running several simulations with the same settings)
- plot_results.R: R script to compare different settings, produces plots and summary table
- plot_results_violin.R: like plot_results.R but TBL are plotted as violin plots
- plot_results_fig3.R: slight variation of plot_results.R to reproduce Fig.3
- plot_results_fig4.R: slight variation of plot_results.R to reproduce Fig.4
- MTB_sim.yml: yml file to create the conda environment to run the pipeline
- R_info.yml: yml file to create the conda environment to run the R scripts
conda env create --file MTB_sim.yml
This will create a conda environment with all the tools needed, with one exception. Beast2 needs to be installed manually (tested with v 2.6.6), make sure to install the package MASTER as well (tested with v6.1.2).
Now open the file MTB_cluster_sim.py with a text editor (this is the wrapper for the pipeline), the beginning of the file looks like this:
from Bio import SeqIO
from ete3 import Tree
import argparse
import os
import csv
import sys
import re
import subprocess
import random
import shutil
BEAST_PATH ="~/software/beast/bin/beast"
#SCRATCH_PATH= "/path/to/scratch"
SCRATCH_PATH= os.getcwd()
You need to edit the path to your beast launching script.
If you want you can specify a path to a folder where the intermediate files will be saved (handy when working on a cluster with a scratch partition). If you do so uncomment and edit the first line starting with SCRATCH_PATH, and comment the line after.
For long term reproducibility the conda environment has been archived and can be dwnoaded here: https://doi.org/10.5281/zenodo.8094976.
Once you have dowloaded the file you can run:
mkdir MTB_sim_env
tar -xzf MTB_sim_env.tar.gz -C MTB_sim_env
source MTB_sim_env/bin/activate
Now that the virtual environment is active you should run conda-unpack
Please not that you still need to install MASTER and Beast2. You also need to to edit the path to your beast launching script as explained above.
usage: MTB_cluster_sim.py [-h] [-l] [-ts] [-br] [-dr] [-sr] [-er] [-sim_n]
[-cr [...]] [-ps_sr [...]] [-ps_sy [...]] [-c]
[-s lineages|time] [-min_mt] [-max_mt] [-t] [-rpt]
[-rrt] [-SNP_t] [-f]
optional arguments:
-h, --help show this help message and exit
-l , --lineages number of infectious individuals, when the simulation
exceed this number it stops
-ts , --time_sampling
number of years of sampling (starting from present and
going backward
-br , --birth_rate transmission rate
-dr , --death_rate death rate
-sr , --sampling_rate
sampling rate
-er , --exposed_rate
rate at which exposed become infectious
-sim_n , --simulation_number
simulation number ID
-cr [ ...], --clock_rate [ ...]
clock rate (nucleotide substitution per site per
year), multiple values possible
-ps_sr [ ...], --post_sim_sampling_rates [ ...]
probability of each strain to be sampled (post
simulation), multiple rates possible at once: eg.
<-ps_sr 1 0.5 0.1
-ps_sy [ ...], --post_sim_sampling_years [ ...]
sample only in these years (post simulation), multiple
scheme possible at once: eg. <-ps_sy 1,2,3 1,3,5>
default (all)
-c, --clean delete all intermediate file, keep only clustering
results and terminal branch lengths (default: False)
-s lineages|time, --stop lineages|time
stop criterion, the MASTER simulation should stop when
reaching a certain number of infectious existing
lineages("lineages"; specified with -l) or after a
certain time ("time", specified with -t)(default =
"lineages")
-min_mt , --min_master_tips
minimum number of tips in the tree output of MASTER to
accept the simulation
-max_mt , --max_master_tips
max number of tips in the tree output of MASTER to
accept the simulation
-t , --time Simulation time for MASTER, to be used with "time" as
stop criterion. The simulation will stop after the
amount of years specified with this option. If the
lineage goes extinct before the simulation is retained
(Default = 10)
-rpt , --raxml_pars_tree
number of starting parsimony trees for raxml
-rrt , --raxml_rand_tree
number of starting random trees for raxml
-SNP_t , --SNP_threshold
clustering will be performed for all values in the
interval 0-SNP_t (Default = 50)
-f , --force Discard simulation if tree height < f (default: 0)
There are two possible stop conditions which are mutually exclusive: -s lineages|time.
lineages: the simulation stops when a number of living infectious individuals is reached (specified with -l). If the lineage goes extinct before the simulation is discarded.
time: the simulation stop after a certain number of years (specified with -t). If the lineage goes extinct before the simulation is retained.
If we want to simulate the same conditions corresponding to sup-population type 2 in the manuscript (Fig. 4)
conda activate MTB_sim_env
python MTB_cluster_sim.py -ts 10 -br 0.9 -dr 0.5 -sr 0.5 -er 1.7 -sim_n 1 -cr 0.00000007 -s time -t 30 -f 10 -min_mt 100 --clean
conda deactivate
Output files (these files are not deleted with --clean):
- .tr.newick: the transmission tree simulated by MASTER
- .newick: the corresponding phylogenetic tree
- .fasta_var: the fasta file with the simulated sequences (only variable positions)
- .raxml_rescaled: the inferred phylogenetic tree (with corrected branch length)
- .cl_r: the clustering rates at different SNP thresholds (from 0 to
SNP_t) - _ldist.csv: the list of terminal branch lengths in SNPs (tip_ID,SNP)
The name of the output files (before extension) is composed by the values of the different parameters:
br_er_dr_sr_l_min_mt-max_mt_ts_t_f_cr_er_ps_sy_ps_sr_sim_n
There is a R script available for post-processing of simulation results, it relies on R.utils.
I have a conda environment for this and for the plotting script:
You might need to run this:
conda config --append channels conda-forge
and then
conda create --name R_info --file R_info.yml
Run multiple simulations with the same settings (will take a few minutes, decrease number of simulations or min_mt for speedier testing):
for i in {1..5}; do python MTB_cluster_sim.py -ts 10 -br 0.9 -dr 0.5 -sr 0.5 -er 1.7 -sim_n $i -cr 0.00000007 -s time -t 30 -min_mt 100 -f 10 --clean ;echo "this is the $i simulation"; done;
You can collect the results of the 5 simulations as follow:
conda activate R_info
cd sim_0.9_1.7_0.5_0.5_0_100-2500_10_30_10/
Rscript ../collect_res.R 0.9_1.7_0.5_0.5_0_100-2500_10_30_10_7e-08_1,2,3,4,5,6,7,8,9,10_1
This will generate 5 files:
0.9_1.7_0.5_0.5_0_100-2500_10_30_10_7e-08_1,2,3,4,5,6,7,8,9,10_1.all_cl_r_concat: clustering rates for all simulations (one sim per line)0.9_1.7_0.5_0.5_0_100-2500_10_30_10_7e-08_1,2,3,4,5,6,7,8,9,10_1.all_ldist_concat.csv: terminal branch lengths for all tips in all simulations (one tip per line)0.9_1.7_0.5_0.5_0_100-2500_10_30_10_7e-08_1,2,3,4,5,6,7,8,9,10_1.tbl_freq.csv: the spectrum of TBL for all tips in all simulations (count and frequency)0.9_1.7_0.5_0.5_0_100-2500_10_30_10_7e-08_1,2,3,4,5,6,7,8,9,10_1.info: some general info and stats0.9_1.7_0.5_0.5_0_100-2500_10_30_10_7e-08_1,2,3,4,5,6,7,8,9,10_1.N_count: the number of tips in each simulated dataset (one sim per line)
There is a R script available for plotting the results of different simulation settings, it uses the following packages: argparser, ggplot2, data.table, ggpubr.
here the help page:
usage: plot_results.R [--] [--help] [--opts OPTS] [-S -S [-f -F [-l -L
[-o -O [-i -I
compare different simulation settings, generate plots and a summary res
table
flags:
-h, --help show this help message and exit
optional arguments:
-x, --opts RDS file containing argument values
-S, -S plot clustering rates for all SNP threshold =< to this
value [default: 5]
-f, -f stem for output files
-l, -l x labels for plots. If spaces in label use quotes(eg. -l
"x label")
-o, -o comma delimited list of labels for each file in the
desired order, eg. -o 1,2,3,4. If there are spaces within
label use quotes
-i, -i colon delimited list of stem name for each simulation
(including path) same order as in -o. Use quotes if
spaces in path
To reproduce Figure 2 (will take a few minutes):
conda activate R_info
cd sim_results
Rscript ../plot_results.R -S 10 -f Figure2 -l "Median latency period (months)" -o "16.6,8.3,4.2" -i \
latency/1.0_0.5_0.5_0.5_0_100-2500_10_30_0_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
latency/1.0_1.0_0.5_0.5_0_100-2500_10_30_0_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
latency/1.0_2.0_0.5_0.5_0_100-2500_10_30_0_8e-08_1,2,3,4,5,6,7,8,9,10_1
To reproduce Figure 3:
conda activate R_info
cd sim_results
Rscript ../../scripts/plot_results_fig3.R -S 7 -f Figure3 -l "Median infectious period (months) - R0" \
-o "17 - 0.9,17 - 1,17 - 1.1,8 - 0.9,8 - 1,8 - 1.1,4 - 0.9,4 - 1,4 - 1.1" -i \
R0/0.45_1.0_0.0_0.5_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/0.5_1.0_0.0_0.5_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/0.55_1.0_0.0_0.5_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/0.9_1.0_0.0_1.0_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/1.0_1.0_0.0_1.0_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/1.1_1.0_0.0_1.0_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/1.8_1.0_0.0_2.0_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/2.0_1.0_0.0_2.0_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1:\
R0/2.2_1.0_0.0_2.0_0_100-2500_10_30_10_8e-08_1,2,3,4,5,6,7,8,9,10_1
To reproduce Figure 4:
conda activate R_info
cd sim_results
Rscript ../plot_results_fig4.R -S 8 -f Figure4 -l "" -o "Type 1,Type 2" -i \
example/1.1_0.7_0.5_0.5_0_100-2500_10_30_10_1e-07_1,2,3,4,5,6,7,8,9,10_1:\
example/0.9_1.7_0.5_0.5_0_100-2500_10_30_10_7e-08_1,2,3,4,5,6,7,8,9,10_1
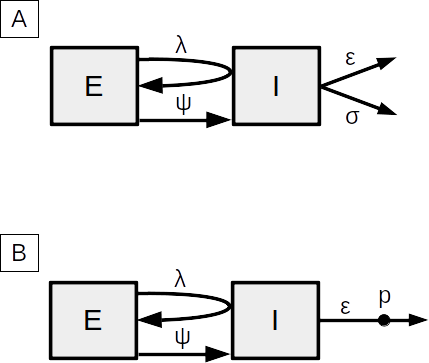
There are two slightly different versions of the BDEI model that can be simulated with this pipeline.
A: This is the model used in the publication. Set the transmission rate (λ)-br, the rate at which exposed individuals become infectious (ψ) -er, the sampling rate (ε) -sr, and the death rate (σ) -dr
B: For this implementation set the death rate -dr to zero and specify the sampling probability (p) with -ps_sr