To predict insertion rates, also check out the MinsePIE online tool on elixir.ut.ee/minsepie/.
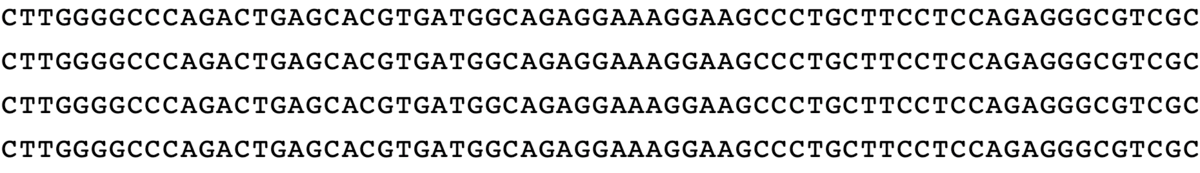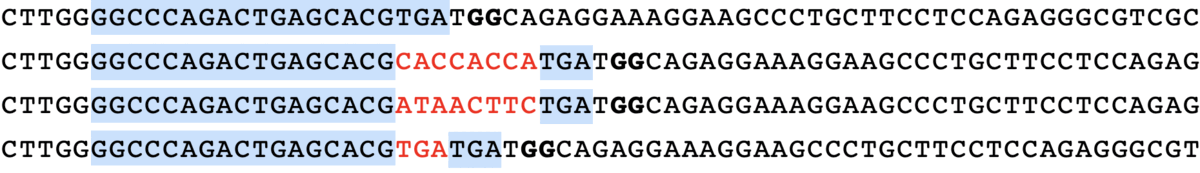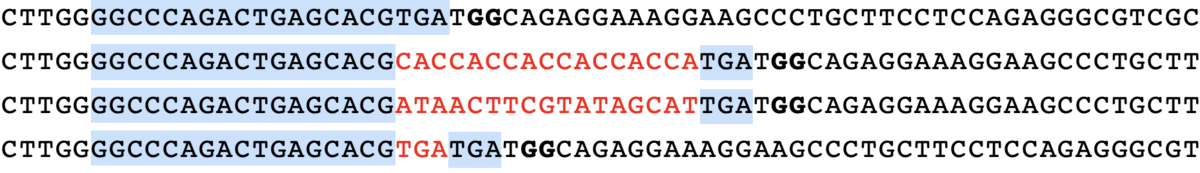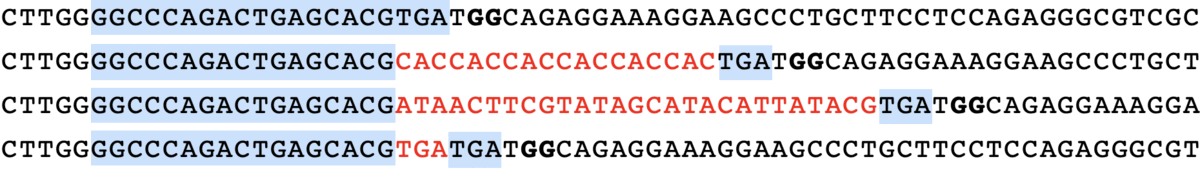
Writing short sequences into the genome with prime eiditng faciliates protein tagging, correction of pathogenic deletions and many more exciting applications. We studied the features that influence insertion efficiency and built a model to predict insertion rates based on the insert sequence. This helps users to choose optimal contructs for DNA insertion with prime editing.
The provided model "MinsePIE.sav" was trained on 22974 events: a libary of 2,666 insert sequences up to 69 nt in length in four genomic sites (CLYBL, EMX1, FANCF, HEK3) in three human cell lines, using the PE2 prime editing system.
System requirements
- Python 3.8
- Python packages: argparse (1.4.0), more_itertools (8.12.0),biopython (1.79), scikit-learn (0.24.2), scipy (1.5.3), XGBoost (1.5.0), pandas (1.3.4), pandarallel (1.5.4), regex (2021.8.3), RNAlib-2.4.18
If you encounter problems setting up the environment or packages, please check out the detailed description for installing the packages in the scripts folder.
The MinsePIE tools are constantly improving. Therefore, it is recommended to run clone the github repository and update it frequently:
# clone
git clone https://github.com/julianeweller/MinsePIE.git
# update
git pull
# install MinsePIE: go into the folder with setup.py
pip install .
Here is an example on how to use minsepie in python:
import minsepie
minsepie.predict(['TGTCA'], pbs = 'CAGACTGAGCACG', ha = 'TGATGGCAGAGGAAAGGAAGCCCTGCTTCCTCCA', spacer = 'GGCCCAGACTGAGCACGTGA', mmr = 0, outdir = "./")
minsepie.predict(insert, fasta = None, pbs = None, ha = None, spacer = None, halen = 15, pbslen = 13, spclen = 20, mmr = 0, inputmode = None, cellline = None, outdir = None, mean = None, std = None, model = None)
Predicts editing outcomes for insert sequences based on pegRNA features given individually or determined from fasta sequence. Provide either fasta (with optionally rttlen, pbslen, spclen) or pbs + rtt + spacer.
| Parameter | Type | Description |
|---|---|---|
| insert | list | Insert sequences to be tested |
| fasta | file | Fasta file with target sequences |
| pbs | str | Primer binding site sequence for the pegRNA |
| ha | str | Homology arm for reverse transcriptase template covering the homology sequence. This does not include the new sequence to be inserted |
| spacer | str | pegRNA spacer |
| halen | int | Length of the RTT. Only needed if target site is provided as fasta. |
| pbslen | int | Length of the PBS. Only needed if target site is provided as fasta. |
| spclen | int | Length of the spacer. Only needed if target site is provided as fasta. |
| mmr | int | Mismatch repair proficiency of cell line. 0: MMR deficient. 1: MMR proficient |
| Inputmode | “dna”, “protein”, or None | Insert sequence can either be nucleotides or amino acids. If none, default is DNA. |
| cellline | str, None | Instead of providing the MMR status directly, cell line can be provided and MMR status is determined based on reference file. |
| outdir | dir | Output directory |
| mean | int, None | Expected mean editing rate for the prime editing screen used to scale the z-factor to an insertion rate. |
| std | int, None | Expected standard deviation for the prime editing screen used to scale the z-factor to an insertion rate. |
| model | str, None | Model used to predict editing efficiency. |
Returns request as dataframe with features and prediction
Example:
predict([“TGTCA”], pbs = “CAGACTGAGCACG”, ha = “TGATGGCAGAGGAAAGGAAGCCCTGCTTCCTCCA”, spacer = “GGCCCAGACTGAGCACGTGA”, mmr = 0)
Prediction of prime editing insertion efficiencies using sequence features and DNA repair determinants
Jonas Koeppel, Juliane Weller, Elin Madli Peets, Ananth Pallaseni, Ivan Kuzmin, Uku Raudvere, Hedi Peterson, Fabio Giuseppe Liberante & Leopold Parts
Nat Biotechnol (2023)
doi: https://doi.org/10.1101/2021.11.10.468024