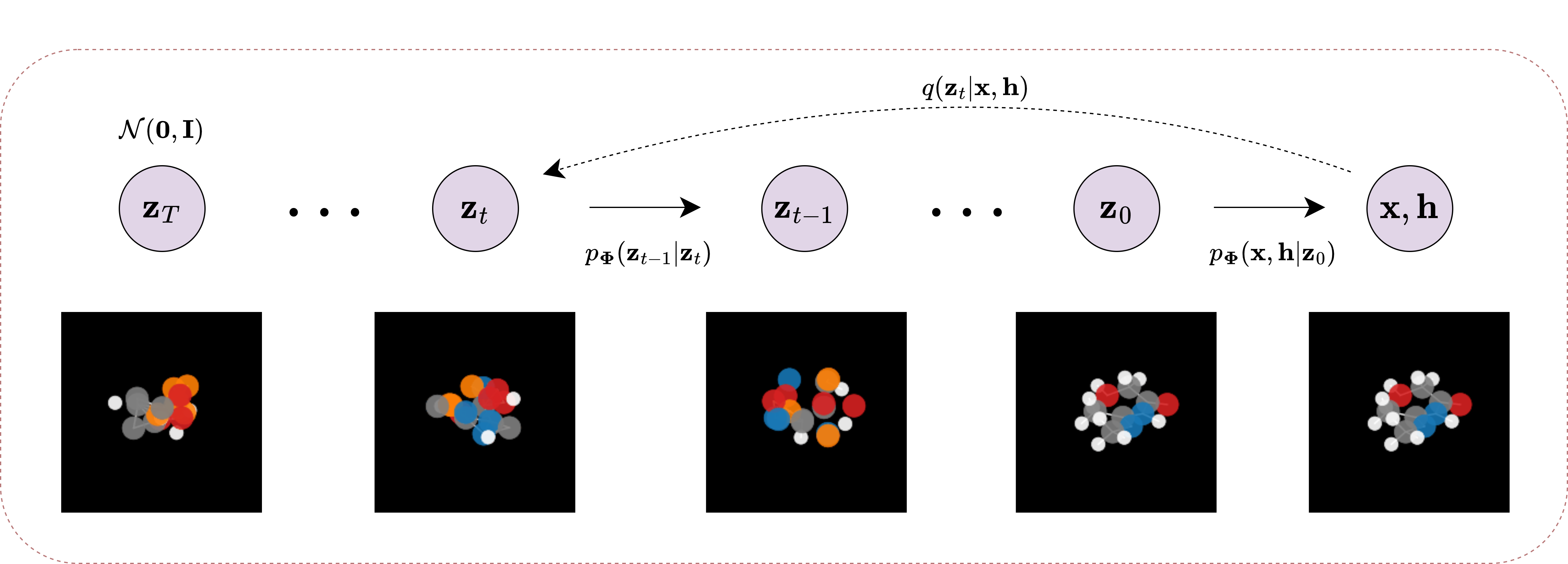
A PyTorch hub of denoising diffusion probabilistic models designed to generate novel biological data
Install Mamba
wget "https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-$(uname)-$(uname -m).sh"
bash Mambaforge-$(uname)-$(uname -m).sh # accept all terms and install to the default location
rm Mambaforge-$(uname)-$(uname -m).sh # (optionally) remove installer after using it
source ~/.bashrc # alternatively, one can restart their shell session to achieve the same resultInstall dependencies
# clone project
git clone https://github.com/BioinfoMachineLearning/bio-diffusion
cd bio-diffusion
# create conda environment
mamba env create -f environment.yaml
conda activate bio-diffusion # note: one still needs to use `conda` to (de)activate environments
# install local project as package
pip3 install -e .Download data
# fetch, extract, and clean-up preprocessed data
wget https://zenodo.org/record/7881981/files/EDM.tar.gz
tar -xzf EDM.tar.gz
rm EDM.tar.gzDownload checkpoints
Note: Make sure to be located in the project's root directory beforehand (e.g., ~/bio-diffusion/)
# fetch and extract model checkpoints directory
wget https://zenodo.org/record/7881986/files/GCDM_Checkpoints.tar.gz
tar -xzf GCDM_Checkpoints.tar.gz
rm GCDM_Checkpoints.tar.gzTrain model with default configuration
# train on CPU
python src/train.py trainer=cpu
# train on GPU
python src/train.py trainer=gpuTrain model with chosen experiment configuration from configs/experiment/
python src/train.py experiment=experiment_name.yamlTrain a model for unconditional small molecule generation with the QM9 dataset (QM9)
python3 src/train.py experiment=qm9_mol_gen_ddpm.yamlTrain a model for property-conditional small molecule generation with the QM9 dataset (QM9)
# choose a value for `model.module_cfg.conditioning` from the properties `[alpha, gap, homo, lumo, mu, Cv]`
python3 src/train.py experiment=qm9_mol_gen_conditional_ddpm.yaml model.module_cfg.conditioning=[alpha]Train a model for unconditional drug-size molecule generation with the GEOM-Drugs dataset (GEOM)
python3 src/train.py experiment=geom_mol_gen_ddpm.yamlNote: You can override any parameter from command line like this
python src/train.py trainer.max_epochs=20 datamodule.dataloader_cfg.batch_size=64Reproduce our results for unconditional small molecule generation with the QM9 dataset
qm9_model_1_ckpt_path="checkpoints/QM9/Unconditional/model_1_epoch_979-EMA.ckpt"
qm9_model_2_ckpt_path="checkpoints/QM9/Unconditional/model_2_epoch_979-EMA.ckpt"
qm9_model_3_ckpt_path="checkpoints/QM9/Unconditional/model_3_epoch_1099-EMA.ckpt"
# note: `trainer.devices=[0]` selects the CUDA device available at index `0` - customize as needed using e.g., `nvidia-smi`
python3 src/mol_gen_eval.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$qm9_model_1_ckpt_path" datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false num_samples=10000 sampling_batch_size=100 num_test_passes=5
python3 src/mol_gen_eval.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$qm9_model_2_ckpt_path" datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false num_samples=10000 sampling_batch_size=100 num_test_passes=5
python3 src/mol_gen_eval.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$qm9_model_3_ckpt_path" datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false num_samples=10000 sampling_batch_size=100 num_test_passes=5Reproduce our results for property-conditional small molecule generation with the QM9 dataset
qm9_alpha_generator_model_filepath="checkpoints/QM9/Conditional/alpha_model_epoch_1619-EMA.ckpt"
qm9_gap_generator_model_filepath="checkpoints/QM9/Conditional/gap_model_epoch_1659-EMA.ckpt"
qm9_homo_generator_model_filepath="checkpoints/QM9/Conditional/homo_model_epoch_1879-EMA.ckpt"
qm9_lumo_generator_model_filepath="checkpoints/QM9/Conditional/lumo_model_epoch_1619-EMA.ckpt"
qm9_mu_generator_model_filepath="checkpoints/QM9/Conditional/mu_model_epoch_1859-EMA.ckpt"
qm9_Cv_generator_model_filepath="checkpoints/QM9/Conditional/Cv_model_epoch_1539-EMA"
qm9_alpha_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_alpha"
qm9_gap_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_gap"
qm9_homo_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_homo"
qm9_lumo_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_lumo"
qm9_mu_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_mu"
qm9_Cv_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_Cv"
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_alpha_generator_model_filepath" classifier_model_dir="$qm9_alpha_classifier_model_dir" property=alpha iterations=100 batch_size=100
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_gap_generator_model_filepath" classifier_model_dir="$qm9_gap_classifier_model_dir" property=gap iterations=100 batch_size=100
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_homo_generator_model_filepath" classifier_model_dir="$qm9_homo_classifier_model_dir" property=homo iterations=100 batch_size=100
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_lumo_generator_model_filepath" classifier_model_dir="$qm9_lumo_classifier_model_dir" property=lumo iterations=100 batch_size=100
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_mu_generator_model_filepath" classifier_model_dir="$qm9_mu_classifier_model_dir" property=mu iterations=100 batch_size=100
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_Cv_generator_model_filepath" classifier_model_dir="$qm9_Cv_classifier_model_dir" property=Cv iterations=100 batch_size=100Reproduce our results for property-specific small molecule optimization with the QM9 dataset
qm9_unconditional_generator_model_filepath="checkpoints/QM9/Unconditional/model_1_epoch_979-EMA.ckpt"
qm9_alpha_conditional_generator_model_filepath="checkpoints/QM9/Conditional/alpha_model_epoch_1619-EMA.ckpt"
qm9_gap_conditional_generator_model_filepath="checkpoints/QM9/Conditional/gap_model_epoch_1659-EMA.ckpt"
qm9_homo_conditional_generator_model_filepath="checkpoints/QM9/Conditional/homo_model_epoch_1879-EMA.ckpt"
qm9_lumo_conditional_generator_model_filepath="checkpoints/QM9/Conditional/lumo_model_epoch_1619-EMA.ckpt"
qm9_mu_conditional_generator_model_filepath="checkpoints/QM9/Conditional/mu_model_epoch_1859-EMA.ckpt"
qm9_Cv_conditional_generator_model_filepath="checkpoints/QM9/Conditional/Cv_model_epoch_1539-EMA.ckpt"
qm9_alpha_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_alpha"
qm9_gap_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_gap"
qm9_homo_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_homo"
qm9_lumo_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_lumo"
qm9_mu_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_mu"
qm9_Cv_classifier_model_dir="checkpoints/QM9/Property_Classifiers/exp_class_Cv"
# unconditionally generate a batch of samples to property-optimize
python3 src/mol_gen_eval_optimization_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false unconditional_generator_model_filepath="$qm9_unconditional_generator_model_filepath" conditional_generator_model_filepath="$qm9_alpha_conditional_generator_model_filepath" classifier_model_dir="$qm9_alpha_classifier_model_dir" num_samples=1000 sampling_output_dir="./optim_mols/" property=alpha iterations=10 num_optimization_timesteps=100 return_frames=1 generate_molecules_only=true use_pregenerated_molecules=false
# optimize generated samples for specific molecular properties
python3 src/mol_gen_eval_optimization_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false unconditional_generator_model_filepath="$qm9_unconditional_generator_model_filepath" conditional_generator_model_filepath="$qm9_alpha_conditional_generator_model_filepath" classifier_model_dir="$qm9_alpha_classifier_model_dir" num_samples=1000 sampling_output_dir="./optim_mols/" property=alpha iterations=10 num_optimization_timesteps=100 return_frames=1 generate_molecules_only=false use_pregenerated_molecules=true
python3 src/mol_gen_eval_optimization_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false unconditional_generator_model_filepath="$qm9_unconditional_generator_model_filepath" conditional_generator_model_filepath="$qm9_gap_conditional_generator_model_filepath" classifier_model_dir="$qm9_gap_classifier_model_dir" num_samples=1000 sampling_output_dir="./optim_mols/" property=gap iterations=10 num_optimization_timesteps=100 return_frames=1 generate_molecules_only=false use_pregenerated_molecules=true
python3 src/mol_gen_eval_optimization_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false unconditional_generator_model_filepath="$qm9_unconditional_generator_model_filepath" conditional_generator_model_filepath="$qm9_homo_conditional_generator_model_filepath" classifier_model_dir="$qm9_homo_classifier_model_dir" num_samples=1000 sampling_output_dir="./optim_mols/" property=homo iterations=10 num_optimization_timesteps=100 return_frames=1 generate_molecules_only=false use_pregenerated_molecules=true
python3 src/mol_gen_eval_optimization_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false unconditional_generator_model_filepath="$qm9_unconditional_generator_model_filepath" conditional_generator_model_filepath="$qm9_lumo_conditional_generator_model_filepath" classifier_model_dir="$qm9_lumo_classifier_model_dir" num_samples=1000 sampling_output_dir="./optim_mols/" property=lumo iterations=10 num_optimization_timesteps=100 return_frames=1 generate_molecules_only=false use_pregenerated_molecules=true
python3 src/mol_gen_eval_optimization_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false unconditional_generator_model_filepath="$qm9_unconditional_generator_model_filepath" conditional_generator_model_filepath="$qm9_mu_conditional_generator_model_filepath" classifier_model_dir="$qm9_mu_classifier_model_dir" num_samples=1000 sampling_output_dir="./optim_mols/" property=mu iterations=10 num_optimization_timesteps=100 return_frames=1 generate_molecules_only=false use_pregenerated_molecules=true
python3 src/mol_gen_eval_optimization_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false unconditional_generator_model_filepath="$qm9_unconditional_generator_model_filepath" conditional_generator_model_filepath="$qm9_Cv_conditional_generator_model_filepath" classifier_model_dir="$qm9_Cv_classifier_model_dir" num_samples=1000 sampling_output_dir="./optim_mols/" property=Cv iterations=10 num_optimization_timesteps=100 return_frames=1 generate_molecules_only=false use_pregenerated_molecules=trueReproduce our results for unconditional drug-size molecule generation with the GEOM-Drugs dataset
geom_model_1_ckpt_path="checkpoints/GEOM/Unconditional/model_1_epoch_76-EMA.ckpt"
geom_model_2_ckpt_path="checkpoints/GEOM/Unconditional/model_2_epoch_79-EMA.ckpt"
geom_model_3_ckpt_path="checkpoints/GEOM/Unconditional/model_3_epoch_78-EMA.ckpt"
python3 src/mol_gen_eval.py datamodule=edm_geom model=geom_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$geom_model_1_ckpt_path" datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false num_samples=10000 sampling_batch_size=100 num_test_passes=5
python3 src/mol_gen_eval.py datamodule=edm_geom model=geom_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$geom_model_2_ckpt_path" datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false num_samples=10000 sampling_batch_size=100 num_test_passes=5
python3 src/mol_gen_eval.py datamodule=edm_geom model=geom_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$geom_model_3_ckpt_path" datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false num_samples=10000 sampling_batch_size=100 num_test_passes=5Unconditionally generate small molecules similar to those contained within the QM9 dataset
qm9_model_ckpt_path="checkpoints/QM9/Unconditional/model_1_epoch_979-EMA.ckpt"
output_dir="./"
my_seed_value=123
python3 src/mol_gen_sample.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$qm9_model_ckpt_path" num_samples=250 num_nodes=19 all_frags=true sanitize=false relax=false num_resamplings=1 jump_length=1 num_timesteps=1000 output_dir="$output_dir" seed="$my_seed_value"Property-conditionally generate small molecules similar to those contained within the QM9 dataset
qm9_alpha_model_ckpt_path="checkpoints/QM9/Conditional/alpha_model_epoch_1619-EMA.ckpt"
qm9_gap_model_ckpt_path="checkpoints/QM9/Conditional/gap_model_epoch_1659-EMA.ckpt"
qm9_homo_model_ckpt_path="checkpoints/QM9/Conditional/homo_model_epoch_1879-EMA.ckpt"
qm9_lumo_model_ckpt_path="checkpoints/QM9/Conditional/lumo_model_epoch_1619-EMA.ckpt"
qm9_mu_model_ckpt_path="checkpoints/QM9/Conditional/mu_model_epoch_1859-EMA.ckpt"
qm9_Cv_model_ckpt_path="checkpoints/QM9/Conditional/Cv_model_epoch_1539-EMA.ckpt"
output_dir="./"
my_seed_value=123
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_alpha_model_ckpt_path" property=alpha iterations=100 batch_size=100 sweep_property_values=true num_sweeps=10 output_dir="$output_dir" seed="$my_seed_value"
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_gap_model_ckpt_path" property=gap iterations=100 batch_size=100 sweep_property_values=true num_sweeps=10 output_dir="$output_dir" seed="$my_seed_value"
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_homo_model_ckpt_path" property=homo iterations=100 batch_size=100 sweep_property_values=true num_sweeps=10 output_dir="$output_dir" seed="$my_seed_value"
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_lumo_model_ckpt_path" property=lumo iterations=100 batch_size=100 sweep_property_values=true num_sweeps=10 output_dir="$output_dir" seed="$my_seed_value"
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_mu_model_ckpt_path" property=mu iterations=100 batch_size=100 sweep_property_values=true num_sweeps=10 output_dir="$output_dir" seed="$my_seed_value"
python3 src/mol_gen_eval_conditional_qm9.py datamodule=edm_qm9 model=qm9_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] datamodule.dataloader_cfg.num_workers=1 model.diffusion_cfg.sample_during_training=false generator_model_filepath="$qm9_Cv_model_ckpt_path" property=Cv iterations=100 batch_size=100 sweep_property_values=true num_sweeps=10 output_dir="$output_dir" seed="$my_seed_value"Unconditionally generate drug-size molecules similar to those contained within the GEOM-Drugs dataset
geom_model_ckpt_path="checkpoints/GEOM/Unconditional/model_2_epoch_79-EMA.ckpt"
output_dir="./"
my_seed_value=123
python3 src/mol_gen_sample.py datamodule=edm_geom model=geom_mol_gen_ddpm logger=csv trainer.accelerator=gpu trainer.devices=[0] ckpt_path="$geom_model_ckpt_path" num_samples=250 num_nodes=44 all_frags=true sanitize=false relax=false num_resamplings=1 jump_length=1 num_timesteps=1000 output_dir="$output_dir" seed="$my_seed_value"Bio-Diffusion builds upon the source code and data from the following projects:
We thank all their contributors and maintainers!
If you use the code or data associated with this package or otherwise find this work useful, please cite:
@inproceedings{morehead2023geometrycomplete,
title={Geometry-Complete Diffusion for 3D Molecule Generation},
author={Alex Morehead and Jianlin Cheng},
booktitle={ICLR 2023 - Machine Learning for Drug Discovery workshop},
year={2023},
url={https://openreview.net/forum?id=X-tLu3OUE-d}
}