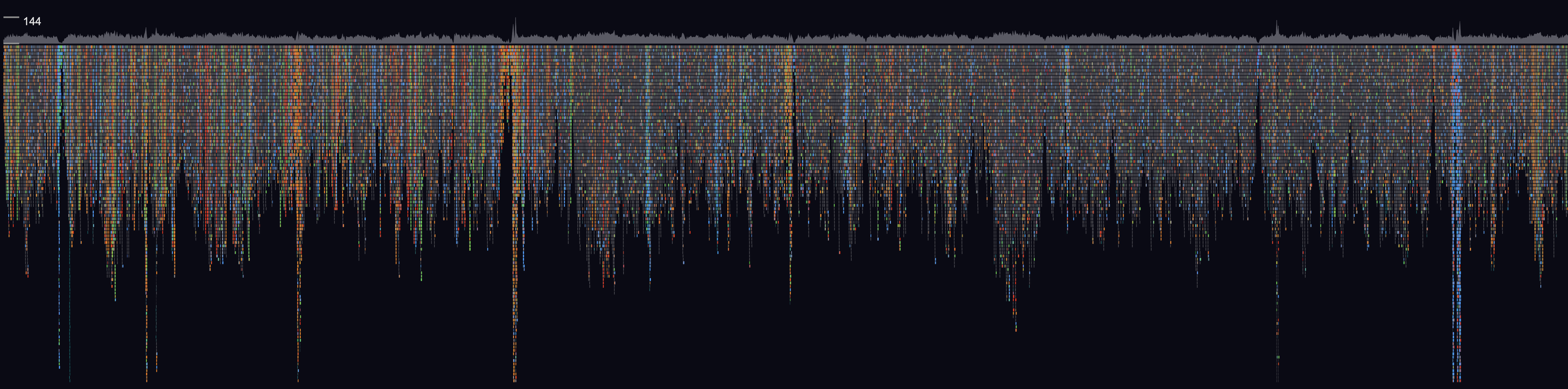
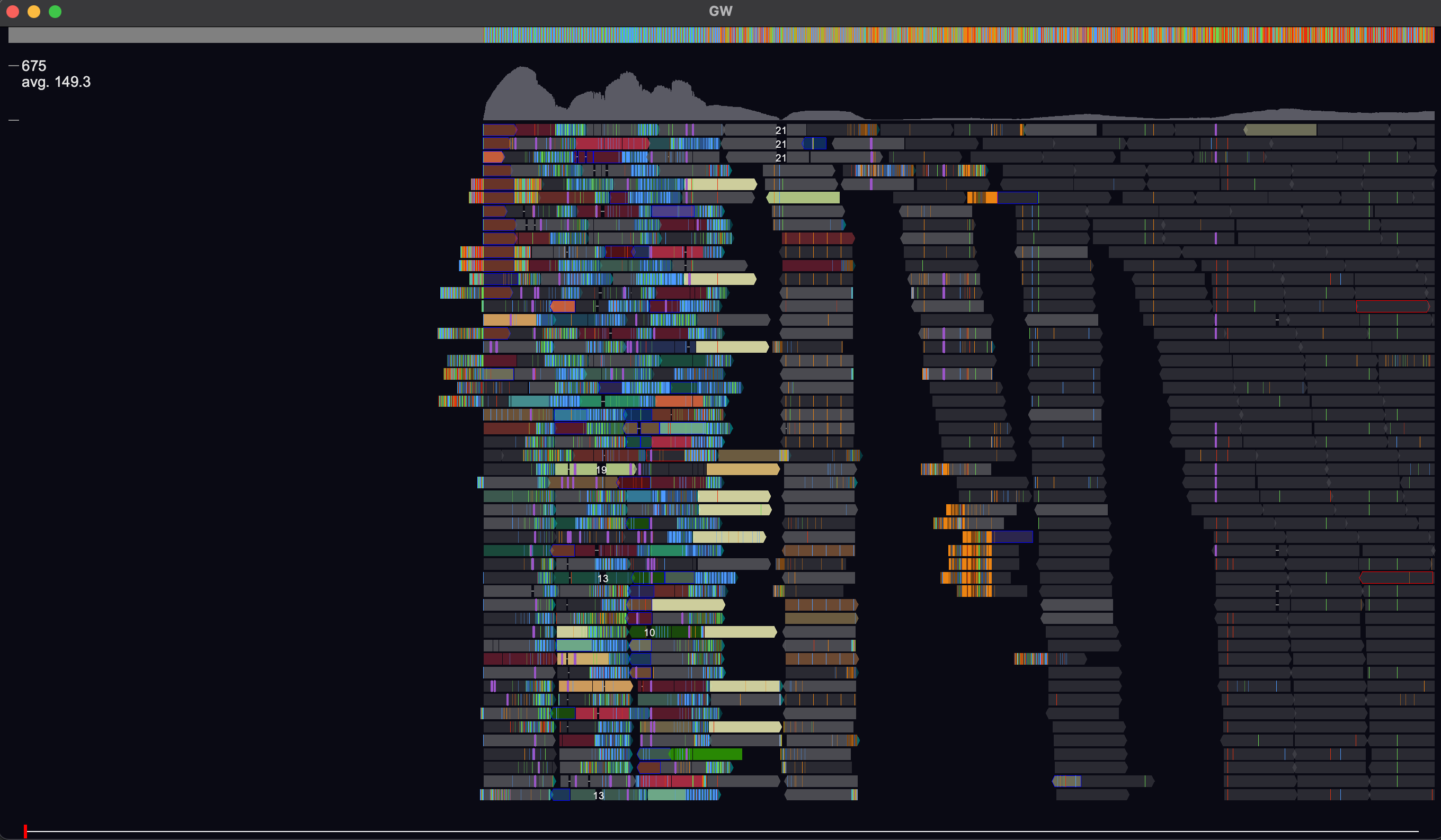
GW is a fast browser for genomic sequencing data (.bam/.cram format) used directly from the terminal. GW also allows you to view and annotate variants from vcf/bcf files.
Linux x86_64 systems:
conda install -c conda-forge -c bioconda gw
Apple x86_64/arm64:
brew install kcleal/homebrew-gw/gw
Windows, install via msys2 or WSL. For msys2, download msys2 then open a ucrt64 terminal (start menu) and install:
pacman -Sy mingw-w64-ucrt-x86_64-gw
Android, install via termux. Head over to the wiki for full instructions!:
pkg install -y gw
A docker container is available with instructions found here:
docker pull kcleal/gw
Building requires glfw3 and htslib libraries:
conda install glfw htslib export CONDA_PREFIX=/pathTo/miniconda # this may already be set git clone https://github.com/kcleal/gw.git && cd gw make prep && make
Build dependencies for ubuntu are also listed in deps folder.
Command line:
# Start gw (drag and drop bams into window) gw hg38 # View start of chr1 gw hg38 -b your.bam -r chr1 # Two regions, side-by-side gw hg38 -b your.bam -r chr1:1-20000 -r chr2:50000-60000 # Multiple bams gw hg38 -b '*.bam' -r chr1 gw hg38 -b b1.bam -b b2.bam -r chr1 # Add a track BED/VCF/BCF/LABEL gw hg38 -b your.bam -r chr1 --track a.bed # png image to stdout gw hg38 -b your.bam -r chr1:1-20000 -n > out.png # Save pdf gw hg38 -b your.bam -r chr1:1-20000 -n --fmt pdf -f out.pdf # plot every chromosome in parallel gw t2t -t 24 -b your.bam -n --outdir chrom_plots # View VCF/BCF gw hg38 -b your.bam -v var.vcf # View VCF/BCF from stdin gw hg38 -b your.bam -v - # View some png images gw -i "images/*.png" # Save some annotations gw hg38 -b your.bam -v var.vcf --labels Yes,No --out-labels labels.tsv
GW commands - access command box with : or /. Example commands:
help # help menu config # open config file for editing chr1:1-20000 # Navigate to region add chr2:1-50000 # Append new region rm 1 # Region at column index 1 removed rm bam1 # Bam file at row index 1 removed mate # Move view to mate of read mate add # mate added in new view line # Toggle vertical line ylim 100 # View depth increased to 100 find QNAME # Highlight all reads with qname==QNAME filter mapq >= 10 # Filer reads for mapq >= 10 count # Counts of all reads for each view point snapshot # Save screenshot to .png man COMMAND # manual for command
To view a genomic region e.g. chr1:1-20000, supply an indexed reference genome and an alignment file (using -b option):
gw hg38 -b your.bam -r chr1:1-20000
The hg38 argument will load a remote reference genome, replace this with the path to a local file for best performance. A GW window will open and can be used interactively with the mouse and keyboard. Note multiple -b and -r options can be used.
Various commands are also available via the GW window. Simply click on the GW window and type help which will display a list of commands in your terminal.
For example typing chr1 will navigate to the start of chromosome 1. For more information about each command type man [command].
A GW window can also be started with only the reference genome as a positional argument:
gw hg38.fa
You can then drag-and-drop alignment files and vcf files into the window, and use commands to navigate to regions etc.
The window size and position can be changed using keyboard keys SHIFT + ARROW_KEY 's.
A variant file in .vcf/.bcf format can be opened in a GW window by either dragging-and-dropping or via the -v option:
gw hg38.fa -b your.bam -v variants.vcf
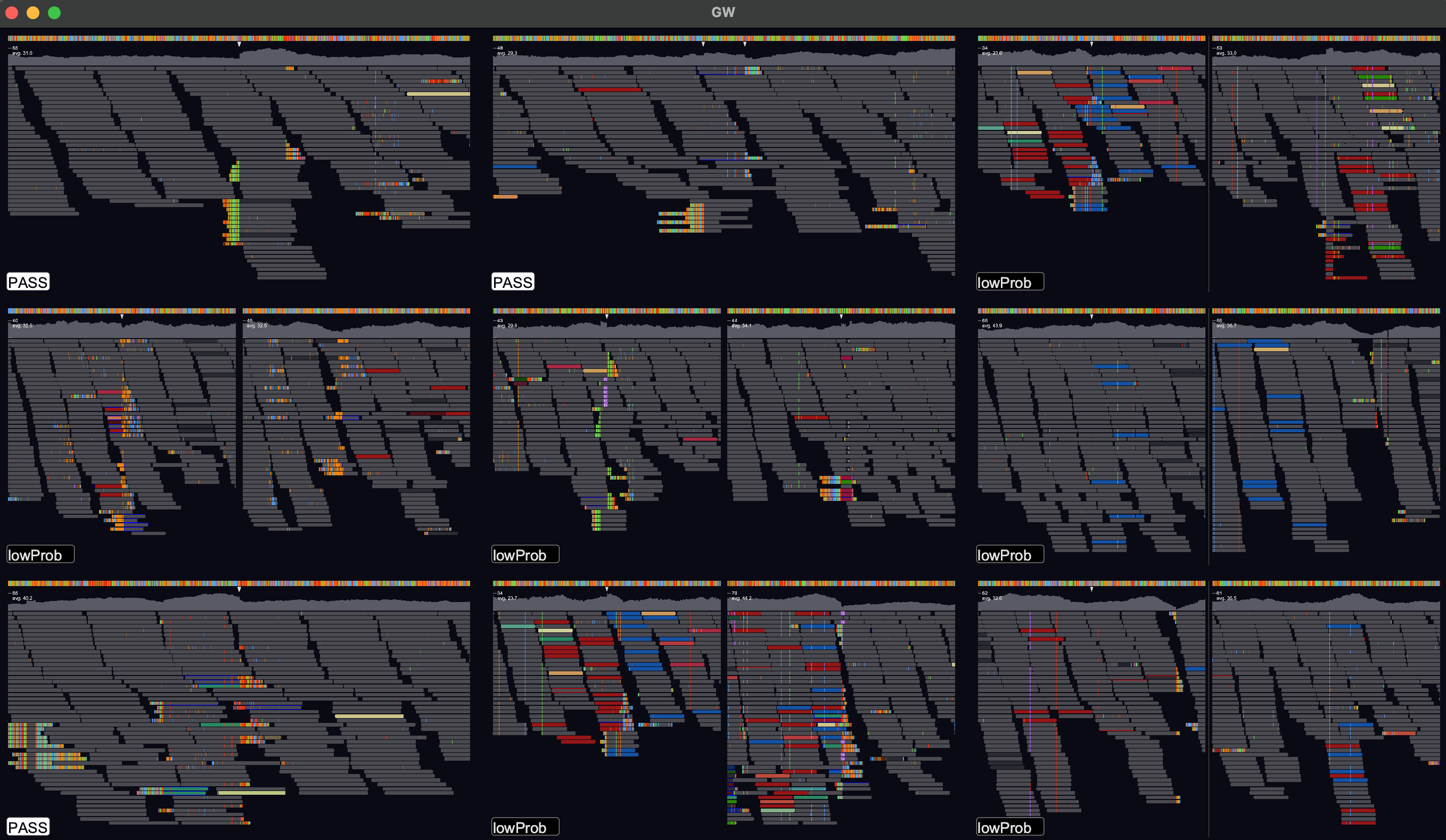
This will open a window in tiled mode. To change the number of tiles use the up/down arrow keys to change interactively or use the -n option to control the dimensions:
gw hg38.fa -n 8x8 -b your.bam -v variants.vcf
If you right-click on one of the tiles then the region will be opened for browsing. To get back to the tiled-image view, just right-click again.
Vcf/bcf files can be open in a stream e.g. using bcftools + gw to select and view regions:
bcftools view -r chr1:1-1000000 your.bcf | gw hg38 -b your.bam -v -
You can also generate an image of every variant in your vcf file - as before use the --outdir and --no-show options. Also,
you might want to increase the number of threads used here to speed things up a bit. Be warned this will probably generate a huge number of files:
gw hg38.fa -b your.bam -v variants.vcf --outdir all_images --no-show -t 16
The time taken here depends a great deal on the speed of your hard drive and depth of coverage, but using a fast NVMe SSD for example, you can expect a throughput around 30-80 images per second.
GW is designed to make manually labelling 100s - 1000s of variants as pain free as possible. Labels can be saved to a tab-separated file and opened at a later date to support labelling over multiple sessions. GW can also write a modified vcf, updating the vcf filter column with curated labels.
To use labelling in GW, first ensure all variant IDs in your input vcf are unique.
When you open a vcf file, GW will parse the 'filter' column and display this as a label in the bottom
left-hand corner of image tiles. Other labels can be parsed from the vcf using the --parse-label option.
For example, the "SU" tag can be parsed from the info column using:
gw hg38.fa -b your.bam -v variants.vcf --parse-label info.SU
Image tiles can then be clicked-on to modify the label, choosing between PASS/FAIL by default.
To provide a list of alternate labels, use the --labels option:
gw hg38.fa -b your.bam -v variants.vcf --labels Yes,No,Maybe
Now when you left-click on a tiled image, you can cycle through this list.
To save or open a list of annotations, we recommend using the --in-labels and --out-labels options. This makes it
straightforward to keep track of labelling progress between sessions. Only variants that have been displayed to screen will be appended to
the results in --out-labels:
gw hg38 -b your.bam -v variants.vcf --in-labels labels.tsv --out-labels labels.tsv
Labels are output as a tab-separated file, for example:
| #chrom | pos | variant_ID | label | var_type | labelled_date |
|---|---|---|---|---|---|
| chr1 | 200000 | 27390 | PASS | DEL | |
| chr1 | 250000 | 2720 | FAIL | SNP | 14-10-2022 16-05-46 |
The labelled_date column is only filled out if one of the tiled images was manually clicked - if this field is blank then
the --parsed-label was used. This feature allows you to keep track of which variants were user-labelled over multiple sessions.
Additionally, the --out-labels file is auto-saved every minute for safe keeping.
GW can also write labels to a vcf file. We recommend using this feature to finalise your annotation - the whole vcf file
will be written to --out-vcf. The final label will appear in the 'filter' column in the vcf. Additionally, the date and previous filter label
are kept in the info column under GW_DATE, GW_PREV:
gw hg38.fa -b your.bam -v variants.vcf --in-labels labels.tsv --out-vcf final_annotations.vcf
Note, the --in-labels option is not required here, but could be used if labelling over multiple sessions, for example. Also,
a GW window will still pop-up here, but this could be supressed using the --no-show option.
Generate images in .png/.pdf format of target genomic regions:
gw hg38 -b your.bam -r chr1:1-20000 -n > out.png gw hg38 -b your.bam -r chr1:1-20000 -n --fmt pdf -f out.pdf
GW can also save the genomic location of a region in the file name, allowing you to open static images alongside bam files. To use this function, first save an image in .png format using GW:
gw hg38.fa -b your.bam -r chr1:1-20000 --outdir . -n
Images saved in .png format can be opened in a similar way to variant data, using the -i or --images option. Files are
input using a glob pattern. For example all .png images in a folder called 'images' can be opened with:
gw -i "images/*.png"
If you previously used GW to generate images from a vcf file (see example in Variant data section), any parsed-labels will be saved in the --outdir directory.
For example if --outdir images was used when generating images, you can now view these images and labels using:
gw -i "images/*.png" --in-labels images/gw.parsed_labels.tsv
To open one or more bam files alongside your images you will need to supply a reference genome. Right-clicking using the mouse will then switch between images and bam files:
gw hg38.fa -b your.bam -i "images/*.png"
To focus on reads of interest, GW can filter reads using simple filter expressions provided via the filter command (or --filter option). The syntax for a filter expression follows "{property} {operation} {value}" (the white-spaces are also needed). For example, here are some useful expressions:
filter mapq >= 20 # only reads with mapping quality >= 20 will be shown filter flag & 2048 # only supplementary alignments are shown filter flag & supplementary # same as above filter ~flag & supplementary # supplementary reads will be removed filter seq contains TTAGGG # Only reads with TTAGGG kmer will be shown filter seq omit AAAAAA # Reads with this kmer will be removed filter mapq > 30 and ~flag & duplicate # also removes duplicate reads filter mapq > 10 or seq-len > 100; ~flag & duplicate # > 1 statements
These expressions will apply filtering to all image panes (regions and bams). If you want to be more selective, you can use array indexing notation to filter on certain rows (bam files) or columns (regions). For example:
filter mapq > 0 [:, 0] # All rows, column 0 (all bams, first region only) filter mapq > 0 [0, :] # Row 0, all columns (the first bam only, all regions) filter mapq > 0 [1, -1] # Row 1, last column
To remove all filters use the refresh command.
Here is the list of properties you can use (see the sam specification for more details on the meaning of tags):
maps, flag, ~flag, name, tlen, abs-tlen, rnext, pos, ref-end, pnext, seq, seq-len, RG, BC, BX, RX, LB, MD, MI, PU, SA, MC, NM, CM, FI, HO, MQ, SM, TC, UQ, AS
These can be combined with operators:
&, ==, !=, >, <, >=, <=, eq, ne, gt, lt, ge, le, contains, omit
Flag properties can be accessed using keywords, for more info see here:
paired, proper-pair, unmapped, munmap, reverse, mreverse, read1, read2, secondary, dup, supplementary
Once reads have been filtered, you can try the count command which will give you an output similar to samtools flagstats. The count command can also be used with an expression e.g.:
count mapq > 0
GW can be used on remote servers by using ssh -X when logging on to the server, a GW window will show up on your local screen. However performance will generally be slow and laggy. We recommend adding an update delay (in miliseconds) using gw --delay 100 ... which can help prevent bandwidth/latency issues.
Alternatively, the screen sharing tool Xpra can offer much better performance for rendering over a remote connecion.
Xpra will need to be installed on local and remote machines. One way to use Xpra is to start GW on port 100 (on remote machine) using:
xpra start :100 --start="gw ref.fa -b your.bam -r chr1:50000-60000" --sharing=yes --daemon=no
You (or potentially multiple users) can view the GW window on your local machine using:
xpra attach ssh:ubuntu@18.234.114.252:100
The :100 indicates the port. If you need to supply more options to the ssh command use e.g. xpra attach ssh:ubuntu@18.234.114.252:102 --ssh "ssh -o IdentitiesOnly=yes -i .ssh/dysgu.pem"
GW ships with a .gw.ini config file. You can manually set various options within the file so you dont have to keep
typing them in every time. The GW command config will open your config file in a text editor for easy access.
Some useful options to set in your .gw.ini file are a list of reference genomes so these can be selected without using a full path. Also things like the theme, image dimensions and hot-keys can be set.
The .gw.ini file can be copied to your home directory or .config directory for safe-keeping - gw will look in these locations before checking the local install directory.
See test directory.
If you find bugs, or have feature requests please open an issue, or drop me an email clealk@cardiff.ac.uk. GW is under active development, and we would welcome any contributions!