PhyloVega
Visualize phylogenetic trees in Vega from Python.
Declarative tree visualizations in Python powered by Vega.
Warning: This package is still under development. Things may still change rapidly.
Declarative Grammar
from phylovega import TreeChart
# Construct Vega Specification
chart = TreeChart.read_newick(
'tree.newick',
height_scale=200,
# Node attributes
node_size=200,
node_color="#ccc",
# Leaf attributes
leaf_labels="id",
# Edge attributes
edge_width=2,
edge_color="#000",
)Interactive trees
Use Vega grammar
How does it work?
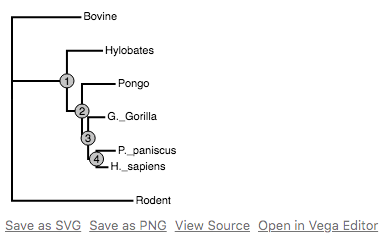
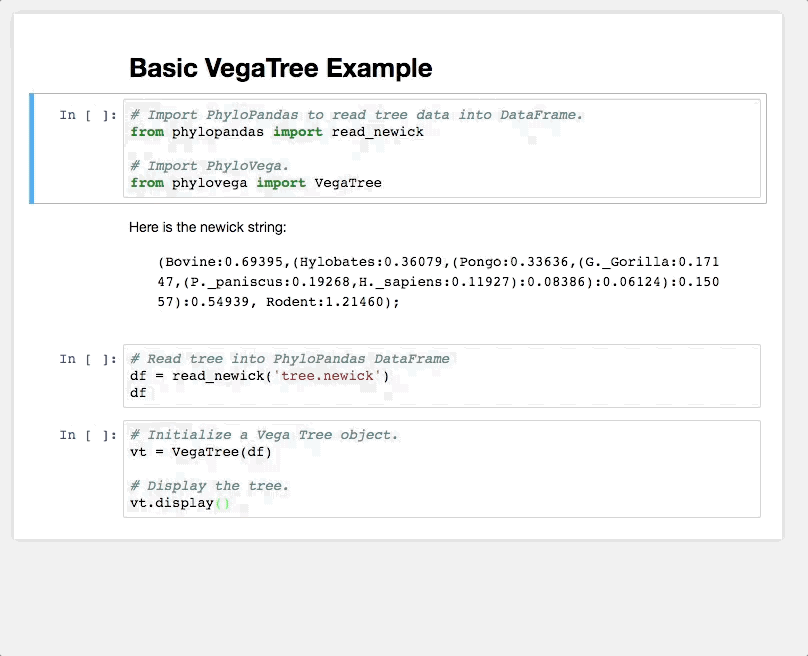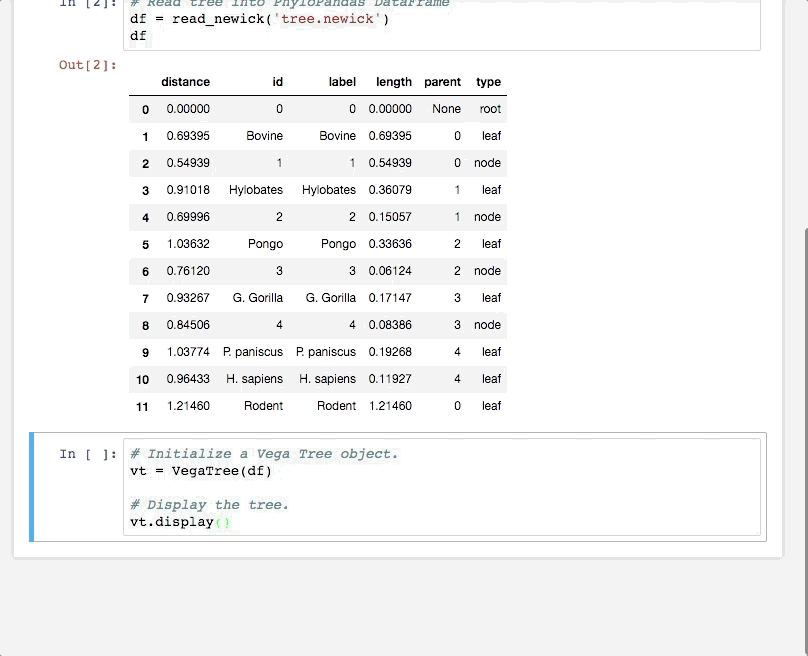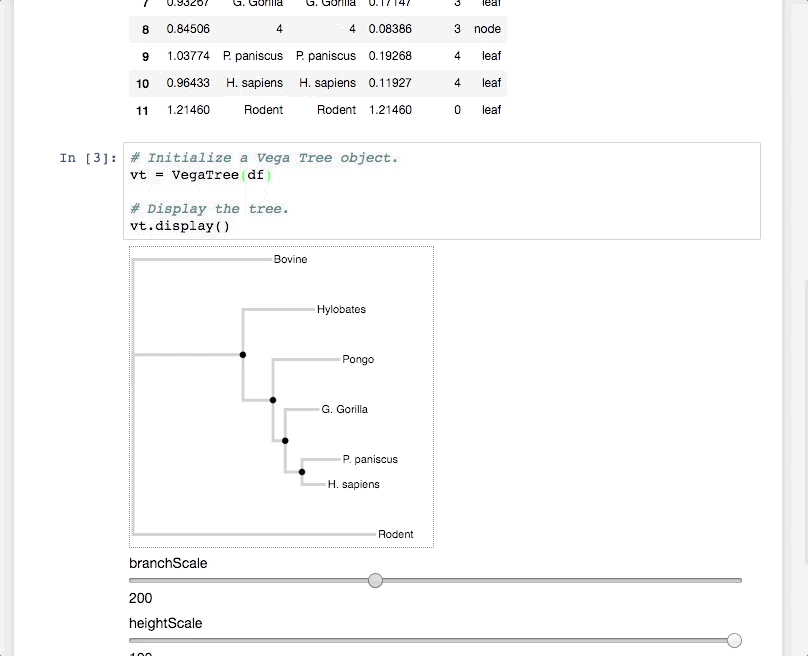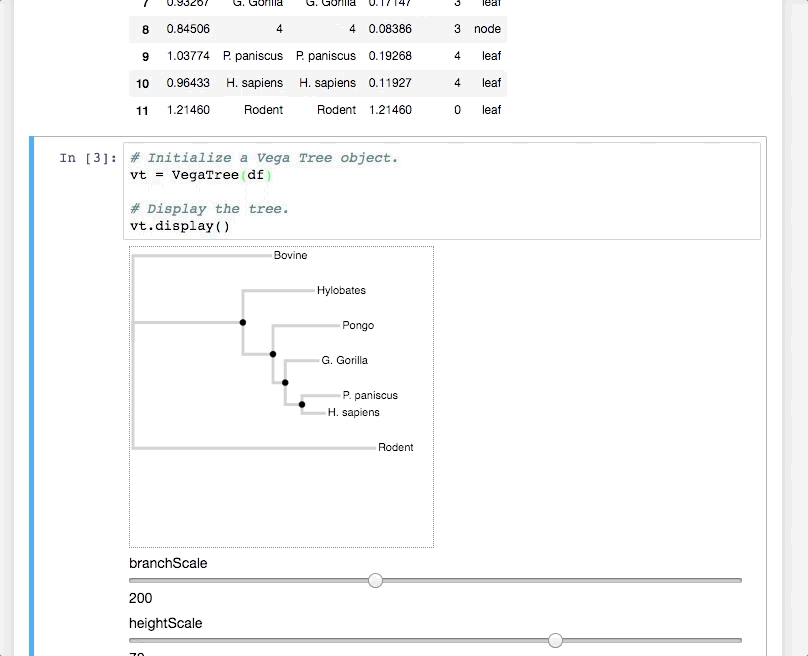
PhyloVega defines uses Vega grammar to draw phylogenetic trees. It accepts a PhyloPandas DataFrame as input and returns a Vega specification in JSON.
Why?
Python is due for a simple, interactive phylogenetic tree viewer. Vega has done most of the heavy lifting here. PhyloVega simply leverages Vega and gets simple interactivity for free!
In the works
Here is a list of features that will eventually make it into PhyloVega.
- Interactive!
- Circular trees
- ... (feel free to add to this list).
Install
Get the latest release with pip:
pip install phylovega
Install the development version by cloning this repo and calling:
pip install -e .
Dependencies
PhyloVega uses the Vega3 specification. To use in the jupyter notebook, you must install the following Python dependencies.
- PhyloPandas: Pandas DataFrame for Phylogenetics
- ipyvega: IPython/Jupyter notebook module for Vega and Vega-Lite (a visualization grammar).