This repository contains code to run the BEEP (Biomedical Evidence Enhanced Predictions) clinical outcome prediction system, described in our NAACL Findings 2022 paper: Literature Augmented Clinical Outcome Prediction
This code was developed in python 3.8 using the libraries listed in environment.yml. The easiest way to run this code is to set up a conda environment using the .yml file via the following command:
conda env create -f environment.yml
Activate the conda environment using the command: conda activate beep-env
After activating the environment, run this command: python -m ipykernel install --user --name beep-env --display-name "Python (beep-env)". This will ensure that beep-env is available as a kernel option when running jupyter notebooks.
In addition to environment setup, you will need access to the MIMIC-III dataset (download here). You will also need to download additional data and trained models from our AWS S3 bucket, especially if you are interested in replicating results from our paper. These resources can be downloaded using the following command:
aws s3 sync --no-sign-request s3://ai2-s2-beep models/
Note that you need to have AWS CLI installed on your machine to execute this command. Move the pubmed_texts_and_dates.pkl file to the data directory.
Our paper evaluates the performance of BEEP on predicting three clinical outcomes:
- Mortality
- Length of Stay
- Prolonged Mechanical Ventilation
For mortality and length of stay, we use the same datasets developed by van Aken et al (2021), which can be obtained here. For mechanical ventilation, the cohort is a subset of the mortality cohort, and the dataset can be constructed by running the following command from the data directory:
python generate_pmv_data <PATH_TO_MORTALITY_DIR>
Note that <PATH_TO_MORTALITY_DIR> refers to the path to the directory containing the train, dev and test files generated for mortality prediction.
To replicate any of our outcome prediction results, you only need to run the outcome prediction module in BEEP, which can be done using the following command:
python run_outcome_prediction.py
--train <PATH_TO_TRAIN_CSV>
--dev <PATH_TO_DEV_CSV>
--test <PATH_TO_TEST_CSV>
--init_model <HF_IDENTIFIER>
--out_dir <PATH_TO_OUTPUT_DIR>
--checkpoint <PATH_TO_TRAINED_MODEL>
--outcome <pmv/los/mortality>
--do_test
--strategy <average/softvote/weightvote/weightaverage>
--lit_dir <PATH_TO_RERANKER_OUTPUT>
--num_top_docs <K_VALUE>
Note that the train, dev and test csv files for every outcome can be generated following the instructions in the previous section on dataset creation.
For the init_model argument, you need to provide the name of the pretrained LM you would like to initialize the model with, as listed on Huggingface or the path to a folder containing a pretrained LM. In our experiments, we primarily use BLUEBERT-Base (bionlp/bluebert_pubmed_mimic_uncased_L-12_H-768_A-12) and UMLSBERT (https://github.com/gmichalo/UmlsBERT).
The checkpoint argument can be used to provide the path to any our trained outcome prediction models, while outcome and strategy can be used to specify which clinical outcome is being predicted and which aggregation strategy is being used (refer to section 3.2.1 for more detail on aggregation strategies).
The lit_dir argument can be used to provide the path to a folder containing ranked abstracts and scores for every EHR, as produced by the cross-encoder reranker which is the final stage of the literature-retrieval module. For better reproducibility, we have included the reranker results for the outcomes and datasets we experiment with under the data folder (e.g., pmv_reranked_results). Finally, the num_top_docs document can be used to specify how many top documents should be used during outcome prediction. Note that the value of this hyperparameter that gives the best results for every setting is included in the trained model name for easy lookup. For example, pmv_bluebert_average_k5.pt indicates that num_top_docs must be set to 5 for this model.
To execute the complete BEEP pipeline on any of our existing datasets (or a new dataset), you will need to run both literature retrieval and outcome prediction. Following is a quick overview of the complete system pipeline:
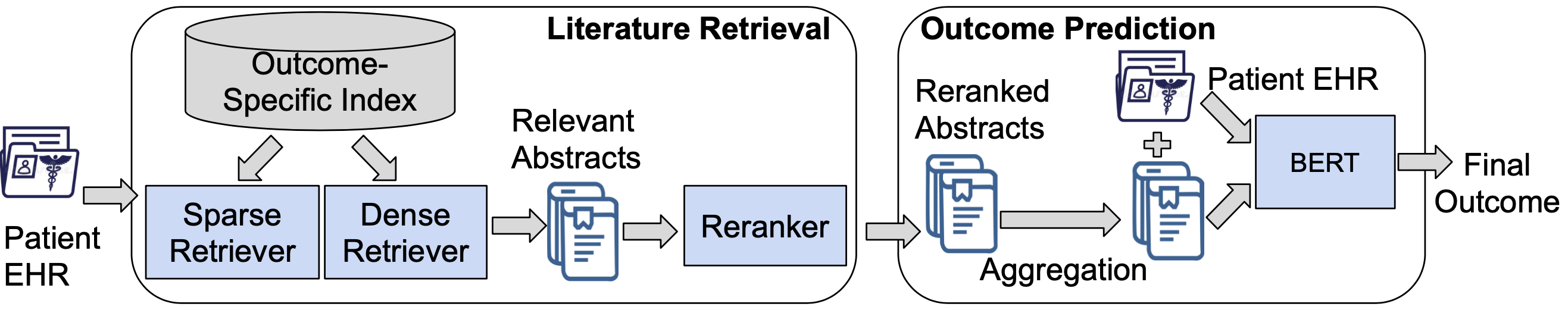
As shown in this pipeline diagram, running BEEP end-to-end consists of five stages:
- Running outcome-specific literature retrieval
- Running sparse retrieval
- Running dense retrieval
- Running cross-encoder reranking
- Running outcome prediction
To retrieve all PubMed literature pertaining to the clinical outcome of interest, run the following notebook present in the literature-retrieval folder: entrez_outcome_specific_retrieval.ipynb.
Note that you will need to make the following changes:
- Add your email ID and tool name to the query URL in cells 2 and 7 to enable entrez API tracking
- Specify additional queries or modify existing queries in cell 3 (currently it contains queries we used to detect mortality literature)
- Change paths to dump output in cells 5 and 9 if needed
Note that running this notebook will query the latest available version of the PubMed database, so you might retrieve a larger set of results than the numbers reported in Table 1(b) even if you use the same set of queries. For better reproducibility, we provide a snapshot of our retrieval results in data/outcome-literature. These files contain the PubMed IDs of articles retrieved for each clinical outcome when we ran the queries (June 2021).
To run sparse retrieval, the first step is to generate MeSH terms for both EHRs and outcome-specific retrieved literature. To perform MeSH term generation from EHRs, first run the mention extractor (in literature-retrieval/mention-extraction) using the following command:
python text_tagger.py
--data <RAW_TEXT_CSV>
--model_name_or_path emilyalsentzer/Bio_Discharge_Summary_BERT
--out_dir <PATH_TO_OUTPUT_DIR>
--checkpoint <PATH_TO_TRAINED_MODEL>
--task i2b2
The input raw text csv file should contain a list of document IDs and corresponding raw texts, while the checkpoint argument can be used to load our trained mention extraction model (located under mention-extraction-models/ in our S3 bucket). Note that our mention extractor is the ClinicalBERT model further finetuned on the i2b2 2010 concept extraction task. If you would like to use a different pretrained language model, or train the mention extractor on a different dataset, we also provide an additional training script (in literature-retrieval/mention-extraction) that can be run as follows:
python pico_trainer.py
--data_dir <DATASET_FOLDER>
--model_name_or_path <HF_IDENTIFIER>
--out_dir <PATH_TO_OUTPUT_DIR>
--task i2b2
--do_train
--do_test
The dataset folder should contain two files corresponding to the train and test splits called train.txt and test.txt (note that a small subset of training data is held out and used for validation because i2b2 2010 did not contain a validation set). These files must follow the CoNLL 2003 format and consist of four space-separate columns. Each word must be placed in a separate line, with the four columns containing the word itself, its POS tag, its syntactic chunk tag, and its named entity tag. After each sentence, there must be an empty line. An example sentence would look as follows:
U.N. NNP I-NP I-ORG
official NN I-NP O
Ekeus NNP I-NP I-PER
heads VBZ I-VP O
for IN I-PP O
Baghdad NNP I-NP I-LOC
. . O O
The model_name_or_path argument can be used to supply a different pretrained model either by providing its string identifier on Huggingface or providing the path to a folder containing the model.
Post extraction, mentions must be processed to remove negated mentions and the filtered mentions are then linked to MeSH concepts. This can be done using the following notebooks in the literature-retrieval folder: mention_filtering.ipynb and mention_linking.ipynb.
Mention filtering requires medspacy which unfortunately requires a version of spacy incompatible with scispacy requirements. Therefore, the filtering notebook alone must be run in a separate environment. To set up this environment, run conda env create -f medspacy_environment.yml and then activate it by running conda activate med-env. After activating the environment, run: python -m ipykernel install --user --name med-env --display-name "Python (med-env)". This will ensure that med-env is available as a kernel option in the notebook. You also need to provide the correct paths to input data (extracted mentions and raw texts) and output data in cells 2 and 5.
Mention linking can be run in the beep-env environment, and requires correct input and output path specification in cells 1 and 3. Note that input to this notebook is the output of the filtering notebook. This MeSH linking procedure is also followed for retrieved literature since MeSH tags present in PubMed do not take term frequency into account. Again for reproducibility, we release final MeSH term outputs for all EHRs and retrieved literature under the mesh-terms folder in our S3 bucket.
Given these MeSH terms for EHRs and outcome-specific literature, the sparse retriever can be run using the following notebook in the literature-retrieval folder: sparse_retriever.ipynb. Again input and output paths can be changed in cells 2 and 7 to point to correct locations.
The dense retrieval model can be run using the following command from literature-retrieval/dense-retriever:
python text_triplet_bireranker.py
--entities <PATH_TO_EHR_MESH_TERMS>
--text_file <PATH_TO_EHR_RAW_TEXTS>
--model_name_or_path microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext
--out_dir <PATH_TO_OUTPUT_DIR>
--checkpoint <PATH_TO_TRAINED_MODEL>
--query_format text
--retrieval_results <PATH_TO_OUTCOME_SPECIFIC_LITERATURE>
--outcome <pmv/los/mortality>
The entities and text_file arguments can be used to provide the paths to files containing MeSH terms and raw texts for EHRs. Note that the dense retriever can use two query formats: (i) text (raw text of EHR note used as query), and (ii) entity (MeSH terms for EHR concatenated and used as query). Our experiments showed that the text format works much better, and is the format used for all experiments and trained models in the final publication.
The checkpoint argument can be used to provide our trained model released under the retrieval-models folder in our S3 bucket. The retrieval_results argument can be used to provide the path to a file containing IDs returned by the outcome-specific retrieval phase. Note that snapshots of our retrieval results are provided in data/outcome-literature.
Our dense retriever is a bi-encoder initialized with the PubMedBERT-Base language model and finetuned on the TREC 2016 literature retrieval task. As with the mention extractor, if you would like to use a different pretrained language model or train on a different dataset, we provide an additional training script, which can be run as follows:
python run_triplet_bireranker_cv.py
--data <PATH_TO_DATASET_FOLDER>
--entities <PATH_TO_MESH_TERMS>
--model_name_or_path <HF_IDENTIFIER>
--out_dir <PATH_TO_OUTPUT_DIR>
--query_format text
--strategy hard
--do_train
--do_test
--years 2016
--folds <NUM_FOLDS>
--epochs <NUM_EPOCHS>
--lr <LEARNING_RATE>
The data argument can be used to provide the path to a folder containing csv files with the train, dev and test data (named train-split-filtered.csv, and so on). These csv files should contain the following columns: example ID, query type (e.g., diagnosis), EHR note text, PubMed article ID, PubMed article text, relevance judgement (0/1). Similar to the previous script, the entities argument can be used to provide MeSH terms for entity-based querying but text-based querying performs better in our experiments. The model_name_or_path argument can be used to supply a different pretrained model either by providing its string identifier on Huggingface or providing the path to a folder containing the model. Setting the folds argument to >1 will run the model in a cross-validation setting, which we did to report performance on TREC since there are no standard train/dev/test splits.
The cross-encoder reranker model can be run using the following command from literature-retrieval/reranker:
python text_reranker.py
--entities <PATH_TO_EHR_MESH_TERMS>
--text_file <PATH_TO_EHR_RAW_TEXTS>
--model_name_or_path microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext
--out_dir <PATH_TO_OUTPUT_DIR>
--checkpoint <PATH_TO_TRAINED_MODEL>
--query_format text
--retrieval_results <PATH_TO_OUTCOME_SPECIFIC_LITERATURE>
--outcome <pmv/los/mortality>
The entities and text_file arguments can be used to provide the paths to files containing MeSH terms and raw texts for EHRs. Similar to the dense retriever, the reranker can also use both entity and text query formats, but the text format works much better, and is used for all final experiments and trained models. The checkpoint argument can be used to provide our trained model released under the retrieval-models folder in our S3 bucket. The retrieval_results argument can be used to provide the path to a folder containing combined results from the dense and sparse retrievers for the outcome of interest. The data/pmv_reranked_final folder gives an example of the format these results must be stored in.
Our reranker is a cross-encoder model also initialized with PubMedBERT-Base and finetuned on TREC 2016. To use a different language model or train on a different dataset, we provide an additional training script, which can be run as follows:
python run_reranker_cv.py
--data <PATH_TO_DATASET_FOLDER>
--entities <PATH_TO_MESH_TERMS>
--model_name_or_path <HF_IDENTIFIER>
--out_dir <PATH_TO_OUTPUT_DIR>
--query_format text
--do_train
--do_test
--years 2016
--folds <NUM_FOLDS>
--epochs <NUM_EPOCHS>
--lr <LEARNING_RATE>
As with the dense retriever training script, the data argument can be used to provide the path to a folder containing csv files with the train, dev and test data (named train-split-filtered.csv, and so on). These csv files should contain the following columns: example ID, query type (e.g., diagnosis), EHR note text, PubMed article ID, PubMed article text, relevance judgement (0/1). The entities argument can be used to provide MeSH terms if you want to try entity-based querying, model_name_or_path can be used to provide a different pretrained model, and folds>1 can be used to run the reranker in a cross-validation setting.
Commands to run the outcome prediction module in test mode are already discussed in detail here. To train an outcome prediction model (with hyperparameter search), run the following command:
python run_outcome_prediction_hpo.py
--train <PATH_TO_TRAIN_CSV>
--dev <PATH_TO_DEV_CSV>
--test <PATH_TO_TEST_CSV>
--init_model <HF_IDENTIFIER>
--out_dir <PATH_TO_OUTPUT_DIR>
--outcome <pmv/los/mortality>
--do_train
--do_test
--strategy <average/softvote/weightvote/weightaverage>
--lit_dir <PATH_TO_RERANKER_OUTPUT>
--epochs <NUM_EPOCHS>
Key changes from the test mode command include: (i) including the do_train flag to run training, (ii) using the epochs argument to specify number of training epochs, (iii) removing the checkpoint argument, and (iv) removing the num_top_docs argument since it is included in the hyperparameter search grid.
If you want to run training for a specific set of hyperparameters instead of running a full grid search, you can call the run_outcome_prediction.py file similarly with the do_train flag and provide your chosen hyperparameter values using arguments num_top_docs, lr, and accumulation_steps.
If you face any issues with the code, models, or with reproducing our results, please contact aakankshan@allenai.org or raise an issue here.
If you find our code useful, please cite the following paper:
@article{naik2021literature,
title={Literature-Augmented Clinical Outcome Prediction},
author={Naik, Aakanksha and Parasa, Sravanthi and Feldman, Sergey and Wang, Lucy Lu and Hope, Tom},
journal={arXiv preprint arXiv:2111.08374},
year={2021}
}