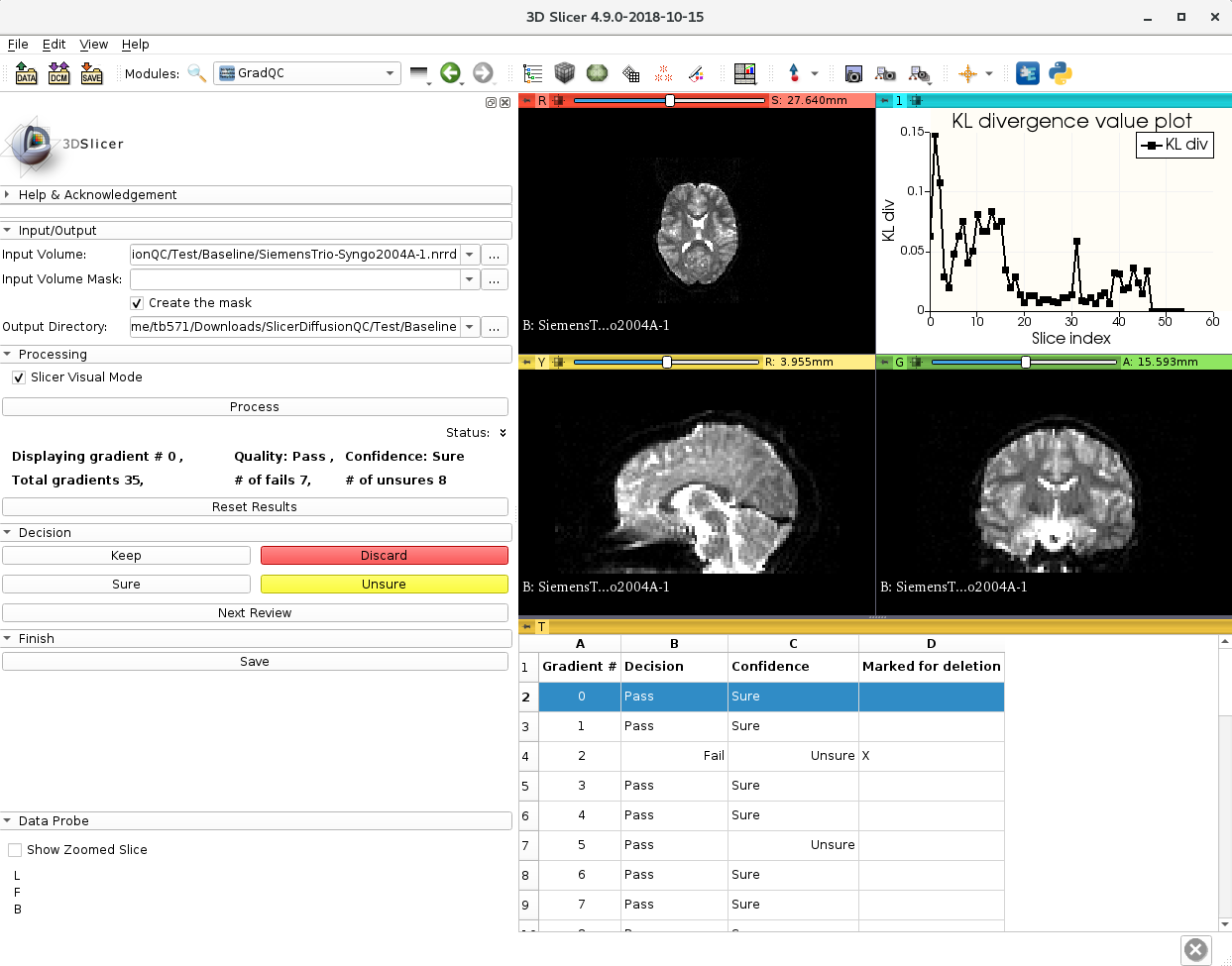
This is a complete slicer module for quality checking of diffusion weighted MRI. It identifies bad gradients by comparing distance of each gradient to a median line. The median line is obtained from KL divergences between consecutive slices. After above processing, it allows user to manually review each gradient: keep or discard them.
A similar software, based on MATLAB environment, was developed earlier by a group of Yogesh Rathi, Asst. Professor, Harvard Medical School. The MATLAB SignalDropQCTool is available at On the other hand, the SlicerDiffusionQC is a faster, cleaner, and more user oriented version of that software.
Developed by Tashrif Billah and Isaiah Norton, Brigham and Women's Hospital (Harvard Medical School).
If you use our software in your research, please cite as below:
Tashrif Billah, Isaiah Norton, Yogesh Rathi, Sylvain Bouix, and Carquex Come, Slicer Diffusion QC Tool, https://github.com/pnlbwh/SlicerDiffusionQC, 2018, Psychiatry Neuroimaging Laboratory, Brigham and Women's Hospital and Harvard Medical School.
System requirement: The program has been extensively tested on Linux Centos7. However, it should run well on any Linux machine. But, we do not have test/support for other platforms (MAC, Windows) yet. We are working on it.
Download Slicer-4.10.0. Slicer Diffusion QC Tool is available as an extension to Slicer. From Extension Manager in Slicer, search DiffusionQC and install.
path/to/slicer.exe --launch python-real /path/to/cli-modules/diffusionQC.py
Checks the quality of gradients in a diffusion weighted mri.
Predicts each gradient as pass or fail.
Usage:
diffusionQC.py [SWITCHES]
Meta-switches:
-h, --help Prints this help message and quits
--help-all Prints help messages of all sub-commands and quits
-v, --version Prints the program's version and quits
Switches:
-a, --auto Turn on this flag for command line/automatic processing w/o Slicer
visualization
-i, --input VALUE:ExistingFile diffusion weighted mri e.g. dwi.nrrd or path/to/dwi.nrrd, accepted formats:
nhdr, nrrd, nii, and nii.gz; required
-m, --mask VALUE:ExistingFile mask for the diffusion weighted mri, e.g. mask.nrrd or path/to/mask.nrrd,
accepted formats: nhdr, nrrd, nii, and nii.gz, if not provided then looks for
default: dwi_mask.format in input directory, if default is not available,
then creates the mask
-o, --out VALUE:str output directory (default: input dwi directory)
i) Download the sample dwi image.
Type GradQC in Slicer>Modules search icon, and select it from the drop down menu. Load the sample dwi, check Create the mask and hit Process.
If the process completes without any error, installation is successfull. You can observe the output files in sample dwi image directory.
ii) From the command line, you can test as follows:
path/to/slicer.exe --launch python-real /path/to/cli-modules/diffusionQC.py -i sample/dwi/image.nrrd -a
ii) (Optional) You can clone the repository and run test as follows:
git clone https://github.com/pnlbwh/SlicerDiffusionQC.git
cd SlicerDiffusionQC
path/to/slicer.exe --launch python-real Test/run_test.py
SlicerDiffusionQC/Test/Baseline contains reference results.
If installation is successful, the above command will create some files in a temporary directory.
Follow STDOUT to see success.
*You need to have git lfs set up on your machine:
Download Git command line extension
tar -xzvf git-lfs-linux-*
PREFIX=$HOME ./install.sh
export PATH=$PATH:$HOME/bin/
git lfs install
The following are example command line calls:
i) If you do not have a mask and you want the pipeline to create one, do not specify the -m/--mask flag.
ii) When you don't have a mask, and you are okay with saving results in the input directory:
path/to/slicer.exe --launch python-real /path/to/cli-modules/diffusionQC.py -i path/to/input.nrrd -a
iii) When you have a mask, and you are okay with saving results in the input directory:
path/to/slicer.exe --launch python-real /path/to/cli-modules/diffusionQC.py -i path/to/input.nrrd -m path/to/mask.nrrd -a
iv) When you want to specify an output diretory:
path/to/slicer.exe --launch python-real /path/to/cli-modules/diffusionQC.py -i path/to/input.nrrd -m path/to/mask.nrrd -o output/directory -a
The above command line calls save temporary .npy files and final results- inputPrefix_modified.nrrd and inputPrefix_QC.csv.
-
Type
GradQCinSlicer>Modulessearch icon, and select it from the drop down menu. -
Specify the input image, input mask, and output directory. Hit
Process. Based on the power of your machine, it might take a few minutes to see the results on Slicer. The rest should be interactive.
The GUI is made in a nice way to load your inputs instantly with mask superimposed on the original image as a labelmap. The Interpolate feature is turned off so you are able to look at the actual signal drops and artifacts.
The CLI wrapper makes following command line calls and creates temporary files as explained below:
path/to/slicer.exe --launch python-real /path/to/cli-modules/diffusionQC.py -i path/to/input.nrrd -m path/to/mask.nrrd
path/to/slicer.exe --launch python-real /path/to/cli-modules/diffusionQC.py -i path/to/input.nrrd -m path/to/mask.nrrd -o output/directory
Saves inputPrefix_QC.npy, inputPrefix_confidence.npy, and inputPrefix_KLdiv.npy. The temporary files are loaded in
Slicer GUI. After user interaction, when Save
is selected, the results are saved as inputPrefix_modified.nrrd and inputPrefix_QC.csv.
If mask is not available, keep the Input Volume Mask field empty and check Create the mask. Then, inputPrefix_bse.nrrd and inputPrefix_mask.nrrd are created.
The following are run modes from GUI:
i) Slicer Visual Mode (checked) and result files exist: does not do gradient processing again.
Uses existing result files to display on Slicer, waits for user interaction and Save.
ii) Slicer Visual Mode (checked) and result files do not exist: runs the whole program, uses created result files to display on Slicer, waits for user interaction and Save.
iii) Slicer Visual Mode (unchecked): irrespective of result files' existence, triggers automode and runs the gradient processing again.
Since autoMode is triggered, Slicer GUI interaction does not open.
- Switching among gradients:
Select each row in the table and the display will update with correspoding gradient.
- Switching among slices:
Select a point in the graph and the axial axis display will update with corresponding slice.
- Sure/Unsure and Pass/Fail:
Click the appropriate pushbutton to classify the current gradient on display. The text in the table will change accordingly. The gradients marked Fail will be discarded when saving as a modified image.
- Next Review:
If you have many gradients and you would want to switch among the unsure predictions only, hit Next Review and the display will update with with next unsure classification.
- Reset Results:
During your modification, if you would like to go back to the machine learning decision, hit this button.
- Save:
Once you are done, Save the results and it will replace any previous modified image with the same prefix in your output directory.
Clone the repository git clone https://github.com/pnlbwh/SlicerDiffusionQC.git in your local directory.
diffusionQC.py is the cli-module that can be run from the command line. On the other hand, GradQC.py is the GUI module that is loaded into Slicer.
The SlicerExecutionModel wrapper describes the input/output of this script for use by Slicer. The Slicer scripted GUI module code is within the SlicerDiffusionQC subdirectory.
Feel free to submit an issue on this github repository. We shall get back to you as early as we can.