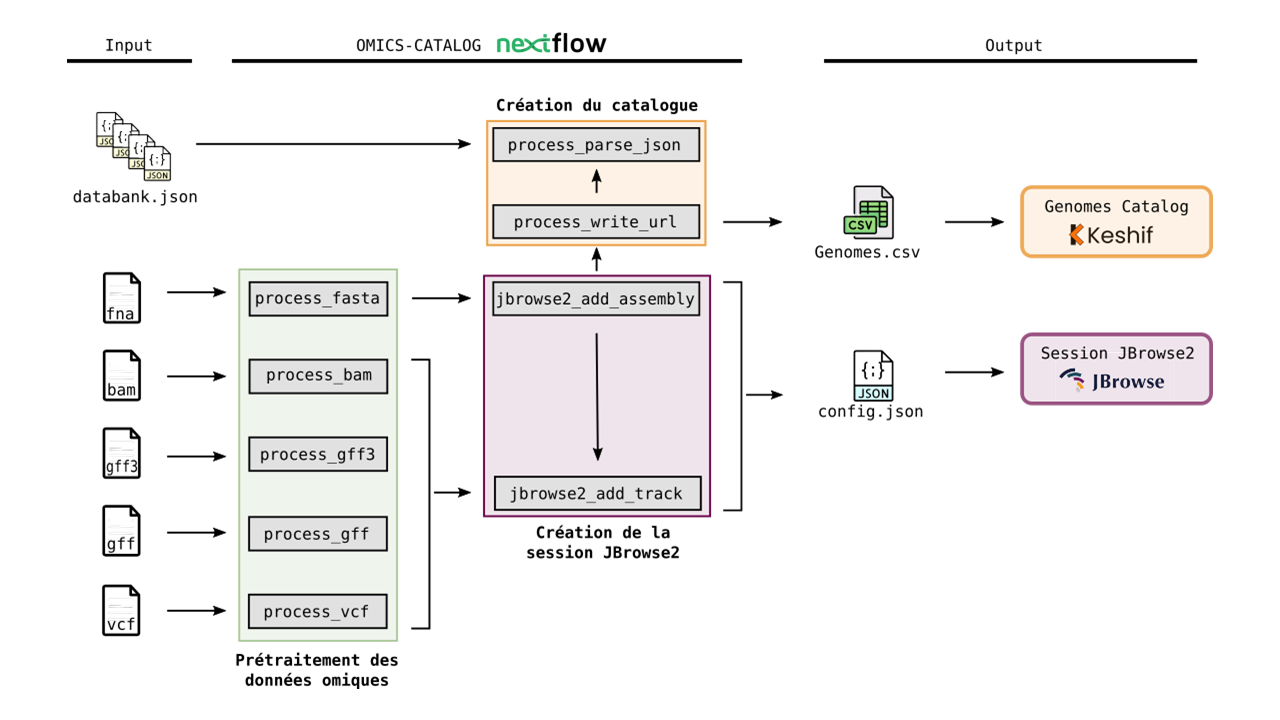
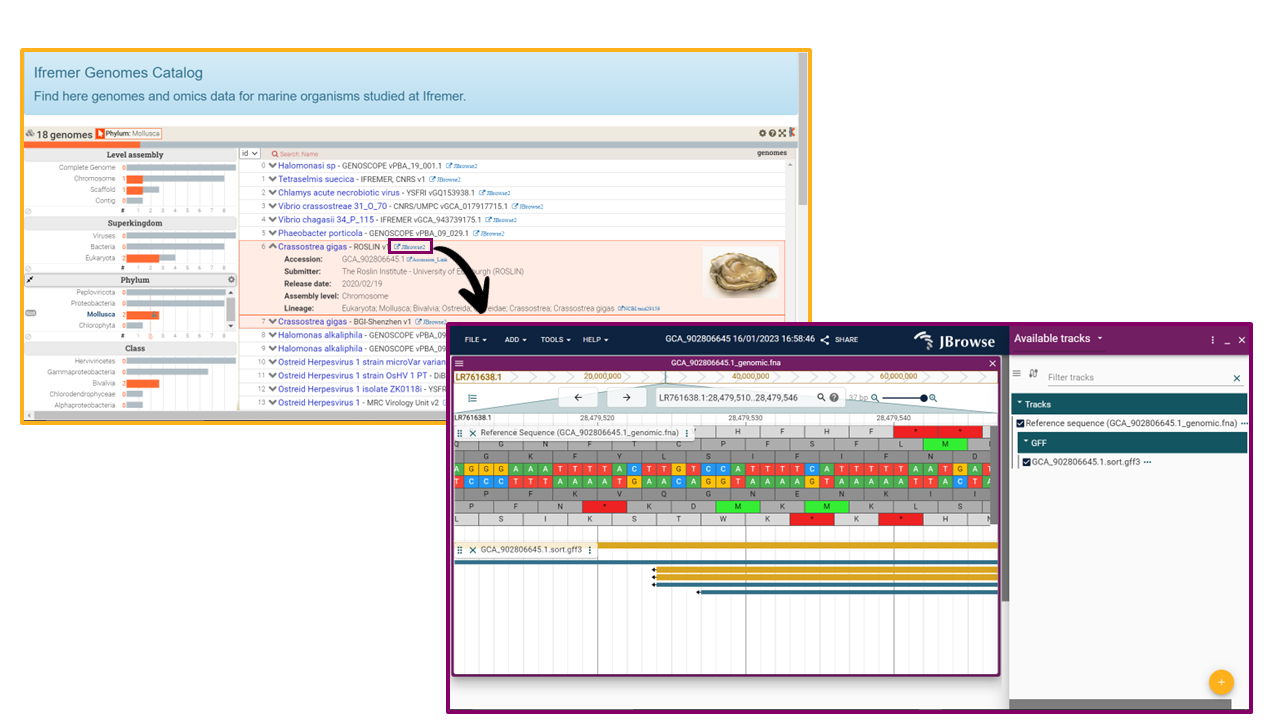
OMICS-CATALOG is based on a Nextflow pipeline which implements a genome browser session to visualize omics data, and sets up an interactive genomes catalog based on the Keshif data visualization tool. The catalog provides the list of available model organisms, along with metadata such as lineage, genome version, submitter, etc. To visualize omics data of model organisms, the catalog gives access to the genome browser session, implemented using JBrowse2 viewer.
The OMICS-CATALOG pipeline can run tasks across multiple compute infrastructures in a very portable manner. It comes with singularity containers making installation trivial and results highly reproducible.
i. Install nextflow
ii. Install Singularity for full pipeline reproducibility
iii. Install JBrowse2
iv. Download the pipeline and test it on a minimal dataset with a single command
cd gviewer
nextflow run main.nf -profile test,singularity -c <institute_config_file>To use this workflow on a computing cluster, it is necessary to provide a configuration file for your system. For some institutes, this one already exists and is referenced on nf-core/configs. If so, you can simply download your institute custom config file and simply use
-c <institute_config_file>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment.
v. Start running your own analysis!
cd gviewer
nextflow run main.nf -profile custom,singularity [-c <institute_config_file>]See usage docs for a complete description of configuration file requirements and all of the options available when running the pipeline.
This workflow comes with documentation about the pipeline, found in the docs/ directory:
- Introduction
- Pipeline installation
- Pre-requisites before running the pipeline
- Running the pipeline
- Output and how to interpret the results
- Troubleshooting
Here is an overview of the many steps available in omics-catalog pipeline:
And here is an oversview of the catalog
To use OMICS-CATALOG, all tools are automatically installed via pre-built singularity images available at SeBiMER ftp; these images are built from recipes available here. The singularity image containing JBrowse2 is used to execute JBrowse2 commands according to the CLI, for setting up the session. However, to access the session you must install JBrowse2 by yourself on your computer. See usage docs for more information.
OMICS-CATALOG is written by SeBiMER, the Bioinformatics Core Facility of IFREMER.
We welcome contributions to the pipeline. If such case you can do one of the following:
- Use issues to submit your questions
- Fork the project, do your developments and submit a pull request
- Contact us (see email below)
For further information or help, don't hesitate to get in touch with the omics-catalog developpers: