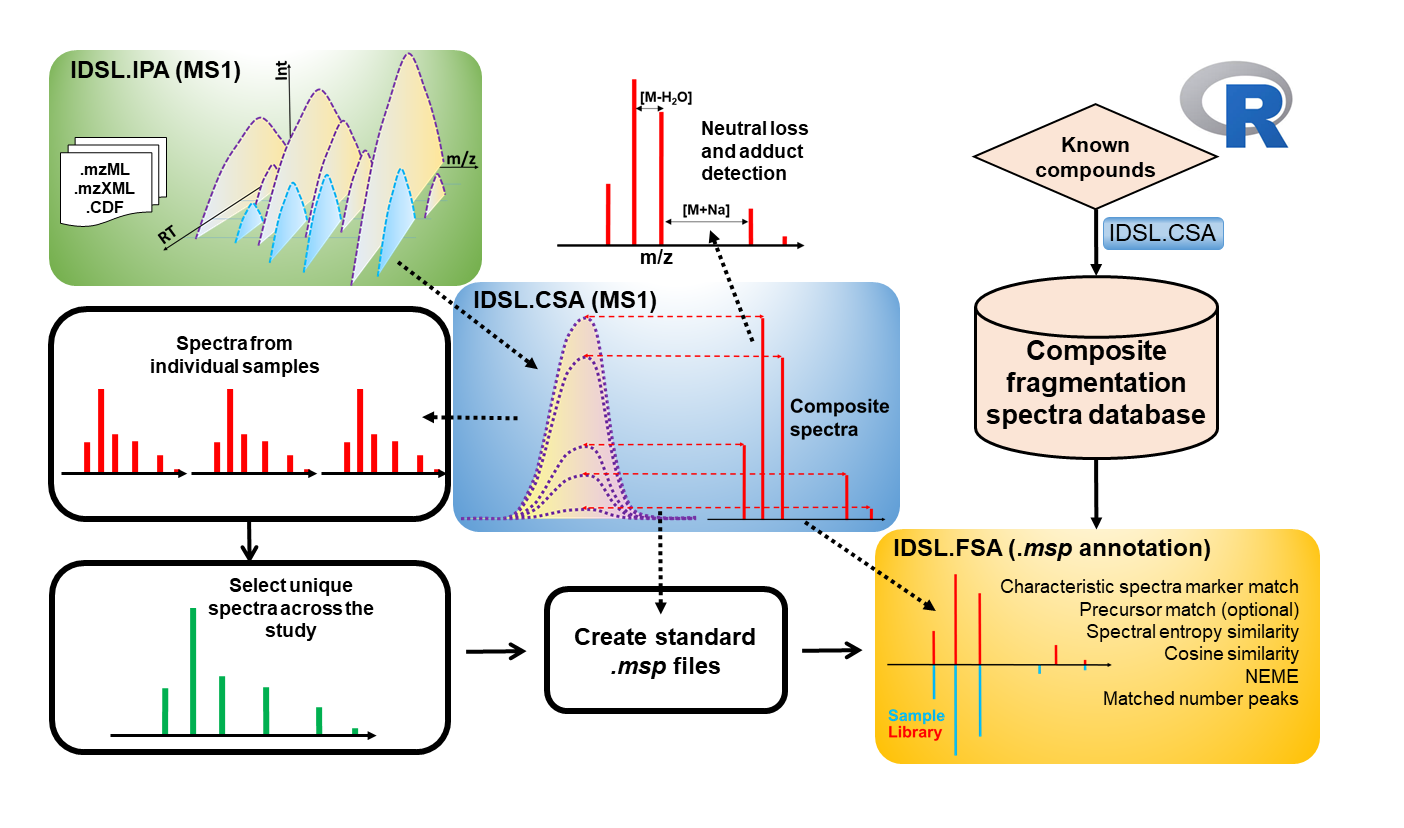
The Composite Spectra Analysis (IDSL.CSA) R package for the analysis of mass spectrometry data has been developed by the Integrated Data Science Laboratory for Metabolomics and Exposomics (IDSL.ME). This package can be used for the deconvolution of fragmentation spectra obtained through various analytical methods such as MS1-only Composite Spectra deconvolution Analysis (CSA), Data Dependent Acquisition (DDA), and a various Data-Independent Acquisition (DIA) methods including MSE, All-Ion Fragmentation (AIF), and SWATH-MS analyses. The aim of the IDSL.CSA package is to assist in streamlining the data analysis process and improving the overall chemical structure annotation in the fields of metabolomics and exposomics.
- Parameter selection through a user-friendly and well-described parameter spreadsheet
- Peak detection and chromatogram deconvolution for various fragmentation data analyses including Composite Spectra Analysis (CSA), Data Dependent Acquisition (DDA), and Data-Independent Acquisition (DIA)
- Analyzing population size untargeted studies (n > 500)
- Aggregating annotated chemical structures on the aligned peak table using meta-variables such as InChIKey, SMILES, precursor type, molecular formula,... depending on the information in the reference library. This is a very unique feature that is only presented by IDSL.CSA. To familiarize with this statistical mass spectrometry feature, try PARAM0006 in the
Starttab in the IDSL.CSA parameter spreadsheet. - Generating batch untargeted aligned extracted ion chromatograms (EIC) figures for the DIA and CSA analyses in addition to generating batch DDA spectra figures.
- Parallel processing in Windows and Linux environments
- Integration with IDSL.FSA workflow to annotate various types of MSP files and generating fragmentation libraries.
install.packages("IDSL.CSA")
Prior to processing your mass spectrometry data (mzXML, mzML, netCDF) using the IDSL.CSA workflow, mass spectrometry data should be processed using the IDSL.IPA workflow to acquire chromatographic information of the peaks (m/z-RT). When the chromatographic information of individual and aggregated aligned peaklists were generated using the IDSL.IPA workflow, download the IDSL.CSA parameter spreadsheet and select the parameters accordingly and then use this spreadsheet as the input for the IDSL.CSA workflow:
library(IDSL.CSA)
IDSL.CSA_workflow("Address of the CSA parameter spreadsheet")
Follow these steps for a quick case study (n = 33) ST002263 which has Thermo Q Exactive HF hybrid Orbitrap data collected in the HILIC-ESI-POS/NEG modes.
-
Process raw mass spectrometry data and chromatographic information using the method described for IDSL.IPA
-
The Composite Spectra Analysis requires 39 parameters distributed into 5 separate sections for a full scale analysis. For this study, use default parameter values presented in the IDSL.CSA parameter spreadsheet. Next, provide information for
2.1. Select YES for PARAM0001 in the
Starttab to only process CSA workflow.2.2. CSA0005 for HRMS data location address (MS1 level HRMS data)
2.3. CSA0008 for Address of the
peaklistsdirectory generated by the IDSL.IPA workflow2.4. CSA0009 for Address of the
peak_alignmentdirectory generated by the IDSL.IPA workflow2.5. CSA0011 for Output location (.msp files and EICs)
2.6. You may also increase the number of processing threads using CSA0004 according to your computational power
-
Run this command in R/Rstudio console or terminal:
library(IDSL.CSA)
IDSL.CSA_workflow("Address of the CSA parameter spreadsheet")
-
You may parse the results at the address you provided for CSA0011.
4.1. CSA_MSP includes .msp file
4.2. CSA_adduct_annotation includes peaklists with potential adduct information
4.3. peak_alignment_subset includes subsets of aligned peak tables for the major ions in each CSA cluster
4.4. aligned_spectra_table includes information for the CSA aggregation on the aligned table
- CSA analysis by IDSL.CSA
- DDA analysis by IDSL.CSA
- DIA analysis by IDSL.CSA
- Unique spectra aggregation
[1] Fakouri Baygi, S., Kumar, Y. Barupal, D.K. IDSL.CSA: Composite Spectra Analysis for Chemical Annotation of Untargeted Metabolomics Datasets. Analytical Chemistry, 2023, 95(25), 9480–9487.
[2] Fakouri Baygi, S., Kumar, Y. Barupal, D.K. IDSL. IPA characterizes the organic chemical space in untargeted LC/HRMS datasets. Journal of proteome research, 2022, 21(6), 1485-1494.