The WFPM package is no longer being actively maintained or developed. ICGC-ARGO is working to transition workflows and packages to utilize the NF-core package manager. This repo will be archived upon completion of full transition.
The WFPM CLI is a command line tool for workflow package authoring and management, it's developed in
Python and runs on a Linux or Mac OS environment. WFPM CLI provides assistance to write shareable/reusable
workflow packages. A package can include one or more of these items: a single step tool
(aka process in Nextflow), a function or a workflow with multiple steps
chained together.
We get a lot of inspiration from NPM (node package manager), which is one of the most successful package mangement systems. The bioinformatics workflow development community would greatly appreciate something like NPM to facilitate and accelerate collaborative development via reusable packages.
NOTE: WFPM CLI is in active development. More features, documentation and tutorials are coming. See Notice on development above.
- Documentation: https://wfpm.readthedocs.io
- Source code: https://github.com/icgc-argo/wfpm
- Talk at BOSC 2021 (including demo): talk; slides.
- Blog: Build workflows collaboratively using reusable and shareable packages
-
Reproducible - same input, same code, same result
- containerize all software tools (including scripts, binary executables) and specific OS environment
- tag every image build and associate it with workflow source code release
-
Portable - run on different platforms, by different users
- containerize all software tools (containerization appeared again, it is a good friend 😊)
- use cross-platform workflow languages and orchestration systems, eg, Nextflow, WDL etc
-
Composable - enable collaborative development
- break down big tasks into small tasks (each carried out by a small software tool)
- one tool per container image
- version and release independently every tool and its associated container image
- a released tool is immutable and can be imported into any workflow where it is needed
- a workflow can also be imported as sub-workflow to build a larger workflow
- similar to tools, workflows are versioned, immutable once released
-
Findable - easy to find by research community members
- register components and workflows in public tool registries, such as Dockstore, BioContainers etc
- release workflow source code via GitHub Releases
-
Testable - deliver with high confidence
- must have tests for every tool, component and workflow
- configure and enable continuous integration testing
Sometime around August 2019, ICGC ARGO started to experiment a modular approach to create workflows using individual analytic tools as reusable building blocks with each tool completely self-contained and independently developed, tested and released. As each tool being fairly small and well decoupled from others, it gave the team high confidence in developing and delivering the tools. Importing a specific version of a tool into a workflow codebase was extremely easy, we were able to reuse same tools in different workflows (residing in different code repositories) for common steps without duplicating a single line of code. In subsequent months, prototyping and testing assured us this was the right approach. Eventually, the aforementioned best practices were established, following which four ICGC ARGO production workflows have been implemented:
- DNA Sequence Alignment Workflow
- Sanger WGS Somatic Variant Calling Workflow
- Sanger WXS Somatic Variant Calling Workflow
- GATK Mutect2 Somatic Variant Calling Workflow
Before having the WFPM CLI tool, a development procedure was followed manually to ensure adherence to the best practices, which was undoubtedly cumbersome and error-prone. Aimed to provide maximized automation and development productivity, the WFPM CLI tool is able to generate templates that include starter workflow code, code for testing, and GitHub Actions code for automated continuous integration (CI) and continuous delivery (CD). We expect WFPM to significantly lower the barriers for scientific workflow developers to adopt the established best practices and accelerate collaborative workflow development within the ICGC ARGO community and beyond.
Please ensure the following prerequisites are met before moving on to installation.
python >= 3.6
pip >= 20.0 (only required for installation)
bash >= 3.2
git >= 2.0
nextflow >= 20.10
docker >= 19.0
pip install wfpm
To update to the latest version, run pip install --upgrade wfpm
To show usage information of WFPM CLI, run wfpm --help, or simply wfpm
We present here step-by-step instructions how to use wfpm to create Nextflow DSL2 workflow packages.
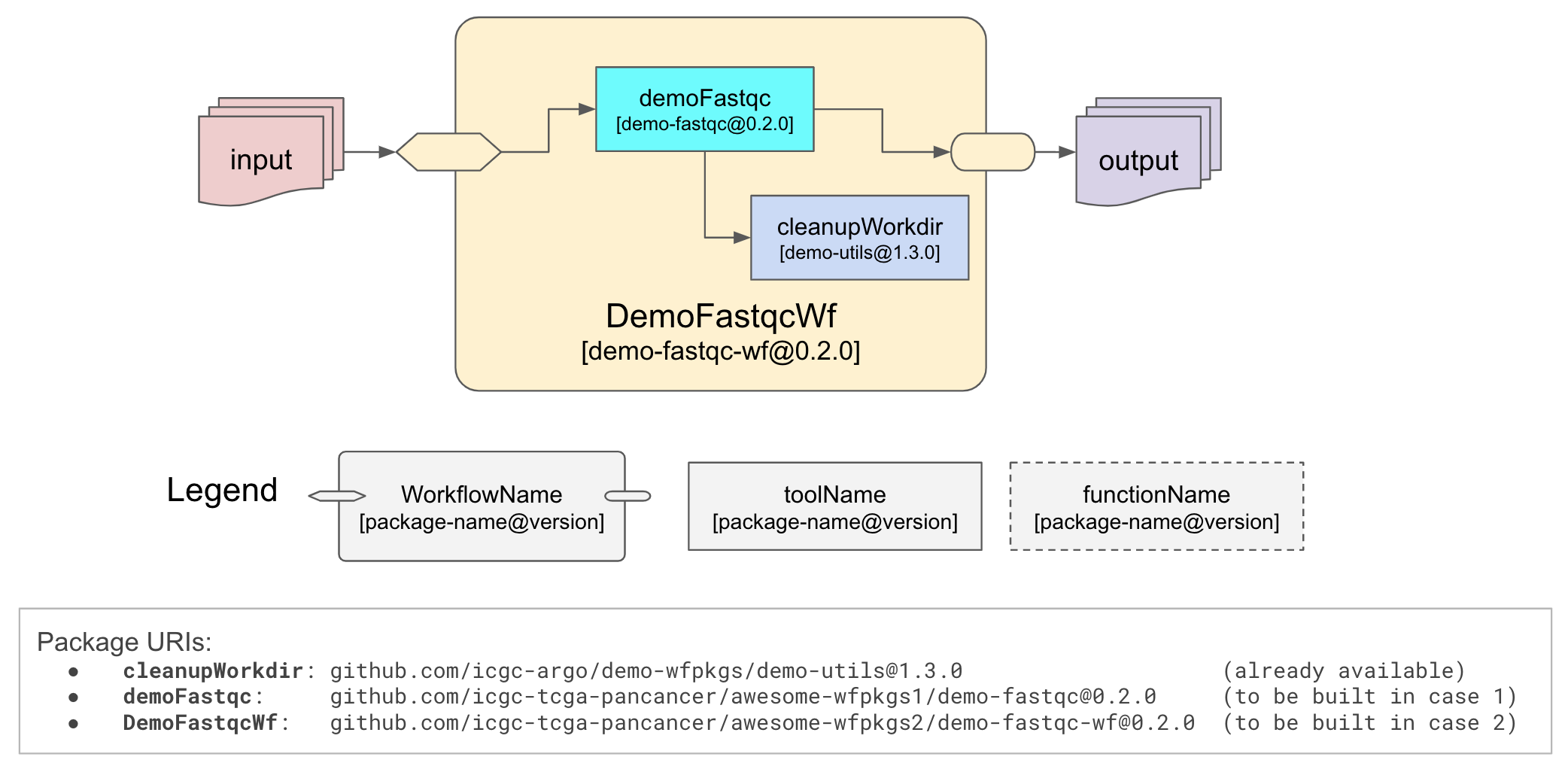
Our objective is to create a workflow that uses FASTQC tool to produce QC metrics for input sequencing
reads. A utility cleanupWorkdir tool is also used to remove unneeded intermediate files. The diagram below
illustrates how the workflow is structured, basically, workflow package demo-fastqc-wf@0.2.0 contains two
tool packages: demo-fastqc@0.2.0 and demo-utils@1.3.0. We will be creating demo-fastqc@0.2.0 and
demo-fastqc-wf@0.2.0 while demo-utils@1.3.0 is already available, we just need to import it as a dependency.
The packages created by the demo cases can be found at: https://github.com/ICGC-TCGA-PanCancer/awesome-wfpkgs1/releases/tag/demo-fastqc.v0.2.0 and https://github.com/ICGC-TCGA-PanCancer/awesome-wfpkgs2/releases/tag/demo-fastqc-wf.v0.2.0 for your reference.
NOTE: You are encouraged to follow these steps to create your own tool / workflow packages. Simply replacing
the GitHub organization ICGC-TCGA-PanCancer used here by your own GitHub account, it should just work.
- Prepare a GitHub repository
Before you start, please make sure you create a repository with name at your choice (in the demo let's
use awesome-wfpkgs1) under a GitHub organization account you have admin access or your personal account
(here we use ICGC-TCGA-PanCancer).
You also need to create a Personal Access Token (PAT) in order to access GitHub Container Registry,
follow these steps: your account => Settings => Developer settings => Personal access tokens =>
Generate new token. Please select write:packages scope for the token.
Once PAT is created, please copy the token and add it to the repository you created above. Here are the
steps to go through: Settings (under the repository page) => Secrets => New repository secret.
For name, please use CR_PAT, value is the PAT you just created.
GitHub Actions greatly helps continuous integration (CI) and continuous delivery (CD) automation.
CI/CD is an integral part of the workflow package development life cycle. To enable GitHub Actions
for your organization: Settings => Actions => Allow all actions. WFPM CLI generated workflow
package templates include all necessary components to perform CI/CD with no work required from you.
- Initialize a project directory for developing/managing packages
wfpm init
Please follow the prompt to provide necessary information. Most important information
includes Project name (this is also the GitHub repo name, please make sure it matches what you have created at step 1. Here we use awesome-wfpkgs1) and GitHub account (we use ICGC-TCGA-PanCancer).
Once completed, you should see something similar as below:
Project initialized in awesome-wfpkgs1
Git repo initialized and first commit done. When ready, you may push to github.com using:
git push -u origin main
When you are ready, as suggested above you can push the code to GitHub. Upon push received at GitHub, CI/CD process will be automatically triggered. You should see CI tests pass, which indicates everything went well.
- Create your first tool package
wfpm new tool demo-fastqc
We use the bioinformatics tool fastqc as an example here. You
can pretty much use the default values in the prompt to advance forward, except for using 0.2.0 for package version. Upon completion,
you should see a message like New package created in: demo-fastqc. Starting template added and committed to git. Please continue working on it. Template code is added to the demo-fastqc@0.2.0 branch,
and WFPM CLI sets the newly created package as currently worked on package, you may verify it by
running:
wfpm workon
You should see the following message:
Packages released: <none>
Packages in development:
demo-fastqc: 0.2.0
Package being worked on: demo-fastqc@0.2.0
When creating your own package, the generated package template gives you the starting point, change the
code as needed. In this demo, the generated demo-fastqc pacakge is already fully functional, we will
just push the code to GitHub:
git push -u origin demo-fastqc@0.2.0
Upon receiving the push, GitHub will automatically start CI/CD via GitHub Actions. If the test
passes, you may create a Pull Request (PR) against the main branch to start the reviewing process.
NOTE: a newly created GitHub container image by default is private, you will need Admin access to make
it public so that anyone is able to pull the image. In this demo case, it can be done on this page:
https://github.com/orgs/ICGC-TCGA-PanCancer/packages/container/awesome-wfpkgs1.demo-fastqc/settings (change the URL
as needed to match your org and repo), click on Change Visibility, then choose Public and confirm.
- Publish your first tool package
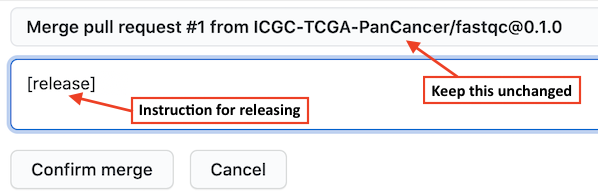
When you merge the above PR, as part of the comment, you may type a special
instruction [release] to let GitHub Actions start the release process, as shown in
the screenshot below. With this GitHub will first merge the demo-fastqc@0.2.0 branch to the main branch,
then starts the release process, once tests are successful, a release of your first tool package
will be made automatically.
The release should be available at: https://github.com/ICGC-TCGA-PanCancer/awesome-wfpkgs1/releases/tag/demo-fastqc.v0.2.0 and can be imported and used by anyone (of course including yourself) in their workflows. How to do that? Please continue to the next demo use case.
In this demo we will be creating a new workflow package that makes use of the demo-fastqc tool package
we created in demo use case 1 (by now it has been released here)
and another utility package published
here: https://github.com/icgc-argo/demo-wfpkgs/releases/tag/demo-utils.v1.3.0
- Prepare another GitHub repository
Similar to the first step of demo use case 1, create another repository (here we use awesome-wfpkgs2)
in the same GitHub organization, add a PAT to it as a secret and name it CR_PAT.
- Initialize a project directory for developing/managing packages
wfpm init
Same as in the previous demo, following the prompt to provide necessary information of the new project.
For Project name and GitHub account, we use awesome-wfpkgs2 and ICGC-TCGA-PanCancer respectively
for this demo.
Upon completion, the scaffold of our second project will be generated and first git commit will be done automatically. You may push the code to GitHub once verified everything is fine.
- Create your first workflow package
Let's name the first workflow package demo-fastqc-wf:
wfpm new workflow demo-fastqc-wf
You may response most of the fields with the default values, except for using 0.2.0 for package version. Notice
that below are dependencies the new workflow requires. Please replace icgc-tcga-pancancer with your own GitHub org
name so the tool package you just released will be used.
github.com/icgc-tcga-pancancer/awesome-wfpkgs1/demo-fastqc@0.2.0github.com/icgc-argo/demo-wfpkgs/demo-utils@1.3.0
wfpm will automatically install and test dependent packages in a temporary directory, once verified
all dependencies tested successfully, they will be copied over to the project space. You should see the
message: New package created in: demo-fastqc-wf. Starting template added and committed to git. Please continue working on it. Template code is added to the demo-fastqc-wf@0.2.0 branch,
and WFPM CLI sets the newly created package as currently worked on package, you may verify it by
running:
wfpm workon
The auto-generated workflow code is fully functional, you may invoke tests as:
wfpm test
This is equivalent to running the tests using Nextflow command directly:
cd demo-fastqc-wf/tests
nextflow run checker.nf -params-file test-job-1.json
nextflow run checker.nf -params-file test-job-2.json
You should see the test run successfully. We now simply push the code to GitHub:
git push -u origin demo-fastqc-wf@0.2.0
CI/CD process will be triggered on the new branch similar to demo 1. Once tests pass, you may create a PR as usual.
- Publish your first workflow package
When merge the PR, type the special instruction [release] in the comment (similar as in the previous demo)
to trigger the CI/CD release process via GitHub Actions. Once released, the demo workflow package will be available at: https://github.com/ICGC-TCGA-PanCancer/awesome-wfpkgs2/releases/tag/demo-fastqc-wf.v0.2.0
By now, you should have a clear picture how WFPM CLI helps to create independent workflow packages and how these packages may be used/reused as building blocks to build larger workflows.
In addition to the packages created by the demo use cases, some more packages are available at: https://github.com/icgc-argo/demo-wfpkgs for your reference.