Stromal_ACTA2_ICI_Analysis
Overview
This repository contains the R-scripts which can reproduce the main figures in the "ACTA2 expression predicts survival and response to immune checkpoint inhibitors in gastric cancer" (submitted to Clinical Cancer Research).
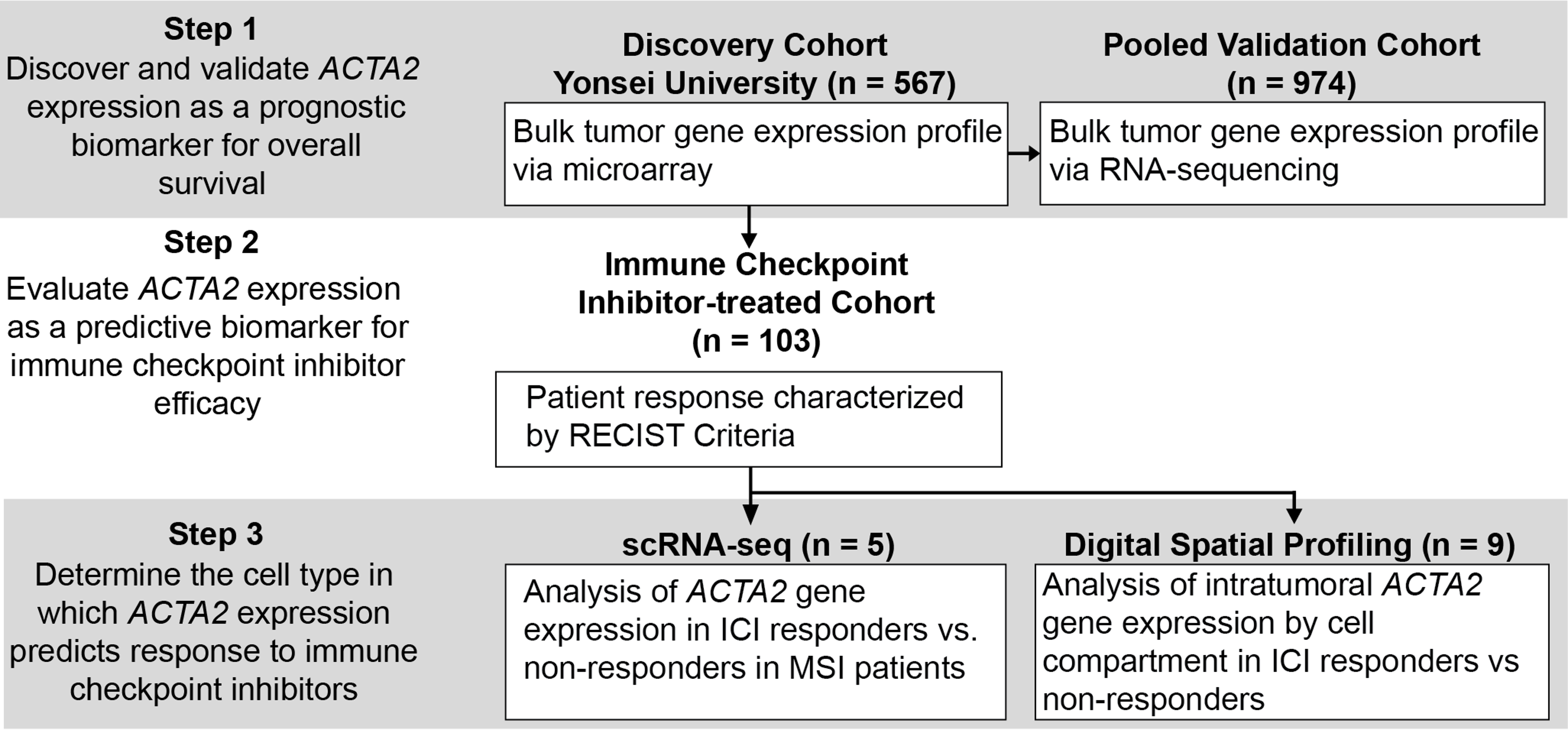
In the paper, we identified the association between tumor ACTA2 expression and overall survival and ICI response in gastric cancer patients. The study workflow is shown in the following figure.
System requirements
The codes require only a standard computer with enough RAM to support the in-memory operations.
OS requirements
The codes are tested on Windows 10.
Installation guide
All the scripts are tested on R (4.1.1). It might take few minutes to complete to install all the required packages.
You can use install.packages("package name") to install the packages.
The required R package: survminer, survival, data.table, ggplot2, ggpubr, plyr, dplyr, and ggbeeswarm
Demo
Please find each script file in "codes" directory and run it in R. The datasets for each script have been included in "data" directory. The figures and additional texts will be generated in "results" directory. The generated figures might show some cosmetic differences from the ones included in the paper as we manually edited the figures for better visualization.
For any questions and comments about the implementations, please contact the authors at park.sunho[nospam]@mayo.edu