This repository provides source code and pre-trained models for fetal brain localization and segmentation from fetal MRI. The method is detailed in [1]. If you use any resources in this repository, please cite the following papers:
-
[1] Michael Ebner*, Guotai Wang*, Wenqi Li, Michael Aertsen, Premal A. Patel, Rosalind Aughwane, Andrew Melbourne, Tom Doel, Anna L. David, Jan Deprest, Sebastien Ourselin, Tom Vercauteren. "An automated localization, segmentation and reconstruction framework for fetal brain MRI." In International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 313-320. 2018. https://doi.org/10.1007/978-3-030-00928-1_36.
-
[2] Eli Gibson*, Wenqi Li*, Carole Sudre, Lucas Fidon, Dzhoshkun I. Shakir, Guotai Wang, Zach Eaton-Rosen, Robert Gray, Tom Doel, Yipeng Hu, Tom Whyntie, Parashkev Nachev, Marc Modat, Dean C. Barratt, Sébastien Ourselin, M. Jorge Cardoso, Tom Vercauteren. "NiftyNet: a deep-learning platform for medical imaging." Computer Methods and Programs in Biomedicine, 158 (2018): 113-122. https://doi.org/10.1016/j.cmpb.2018.01.025.
-
'*' authors contributed equally.
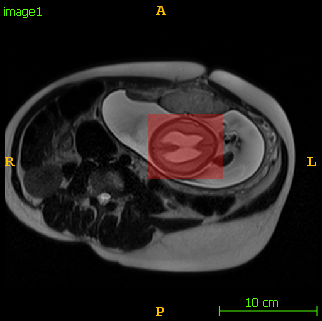
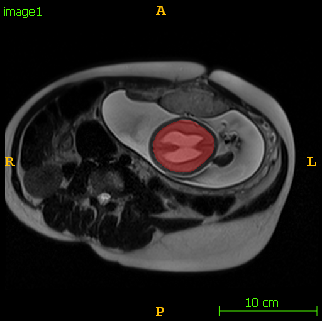
The following images show an example of detection and segmentation results.
For image reconstruction code, please refer to https://github.com/gift-surg/NiftyMIC.
-
A CUDA compatable GPU with memoery not less than 6GB is recommended for training. For testing only, a CUDA compatable GPU may not be required.
-
Tensorflow 1.12.0. Install it by typing
pip install tensorflow-gpu=1.12or following instructions from https://www.tensorflow.org/install. -
NiftyNet 0.2.0. Install it by typing
pip install niftynet=0.2or downloading the source files from https://github.com/NifTK/NiftyNet/releases/tag/v0.2.0. -
Demic (a tool to use NiftyNet) v0.1. Download the source files from https://github.com/taigw/Demic/releases/tag/v0.1 and add the path of Demic to PYTHONPATH environment variable.
-
To get fetal brain detection and segmentation results, run
bash/inference.sh. You need to edit thePYTHONPATHenvironment variable in that file so that it includes the path of NiftyNet and Demic. -
You can edit
cfg_data.txtto customize the input and output image names. -
Alternatively, you can run the following command after setting
PYTHONPATHenvironment variable correctly.
python fetal_brain_seg.py --input_names demo_data/image1.nii.gz demo_data/image2.nii.gz --segment_output_names demo_data/image1_segment.nii.gz demo_data/image2_segment.nii.gz Or the following command that will save detection results.
python fetal_brain_seg.py --input_names demo_data/image1.nii.gz demo_data/image2.nii.gz --segment_output_names demo_data/image1_segment.nii.gz demo_data/image2_segment.nii.gz --detect_output_names demo_data/image1_detect.nii.gz demo_data/image2_detect.nii.gzThis work is part of the GIFT-Surg project (https://www.gift-surg.ac.uk/). It is supported by Wellcome Trust [WT101957; 203145Z/16/Z], EPSRC [EP/L016478/1; NS/A000027/1; NS/A000050/1], and the NIHR UCLH BRC.