Scale‐ and Slice‐aware Net for 3D segmentation of organs and musculoskeletal structures in pelvic MRI
The offical implementation of the network architecture in Paper: Scale- and Slice- aware Net for 3D segmentation of organs and musculoskeletal structures in pelvic MRI
Research Type:Machine Learning/Deep Learning,Image processing/Image analysis, Technical Research
Research Focus:Anatomy, Muscle,Musculoskeletal
Abstract
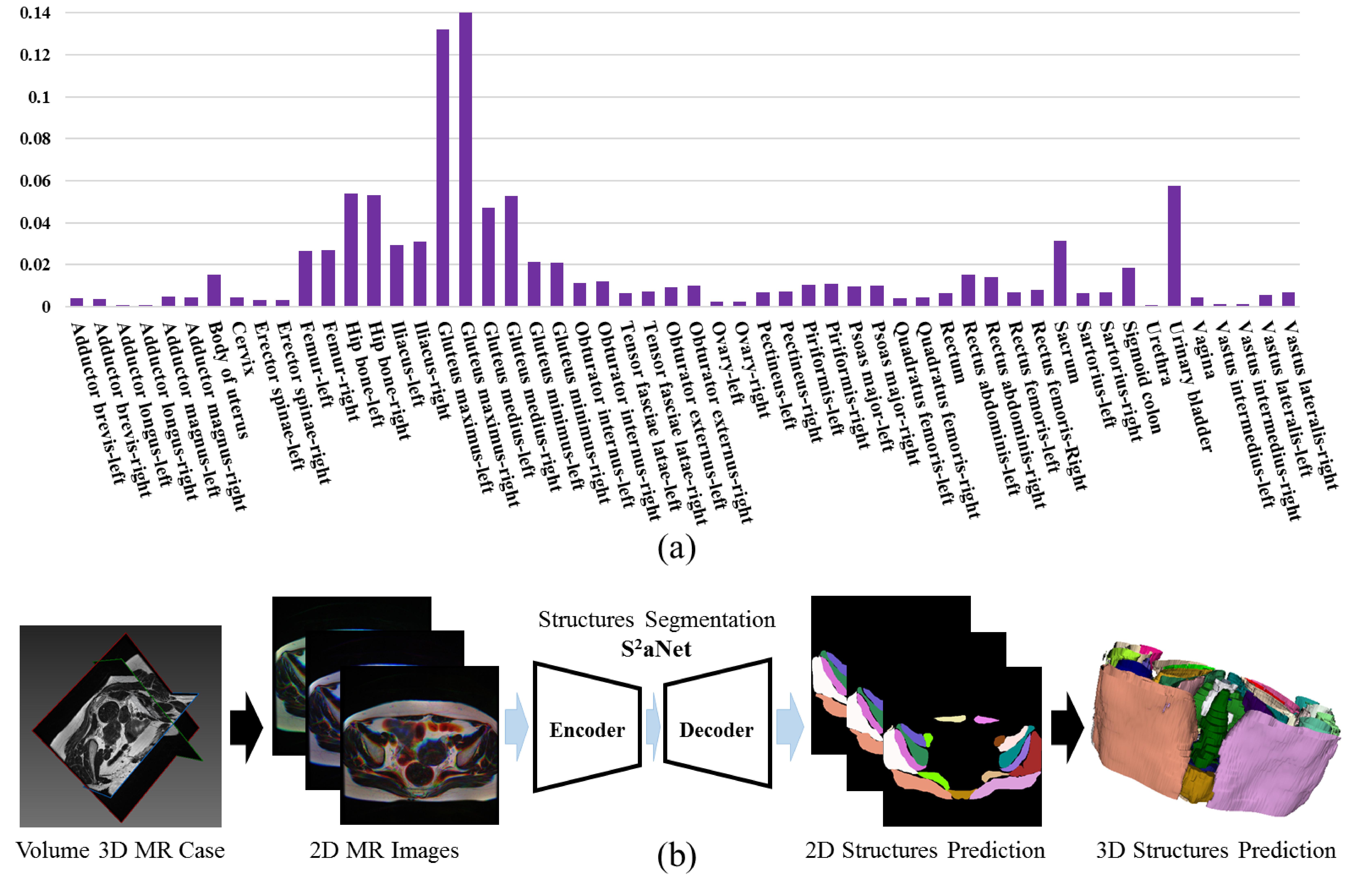
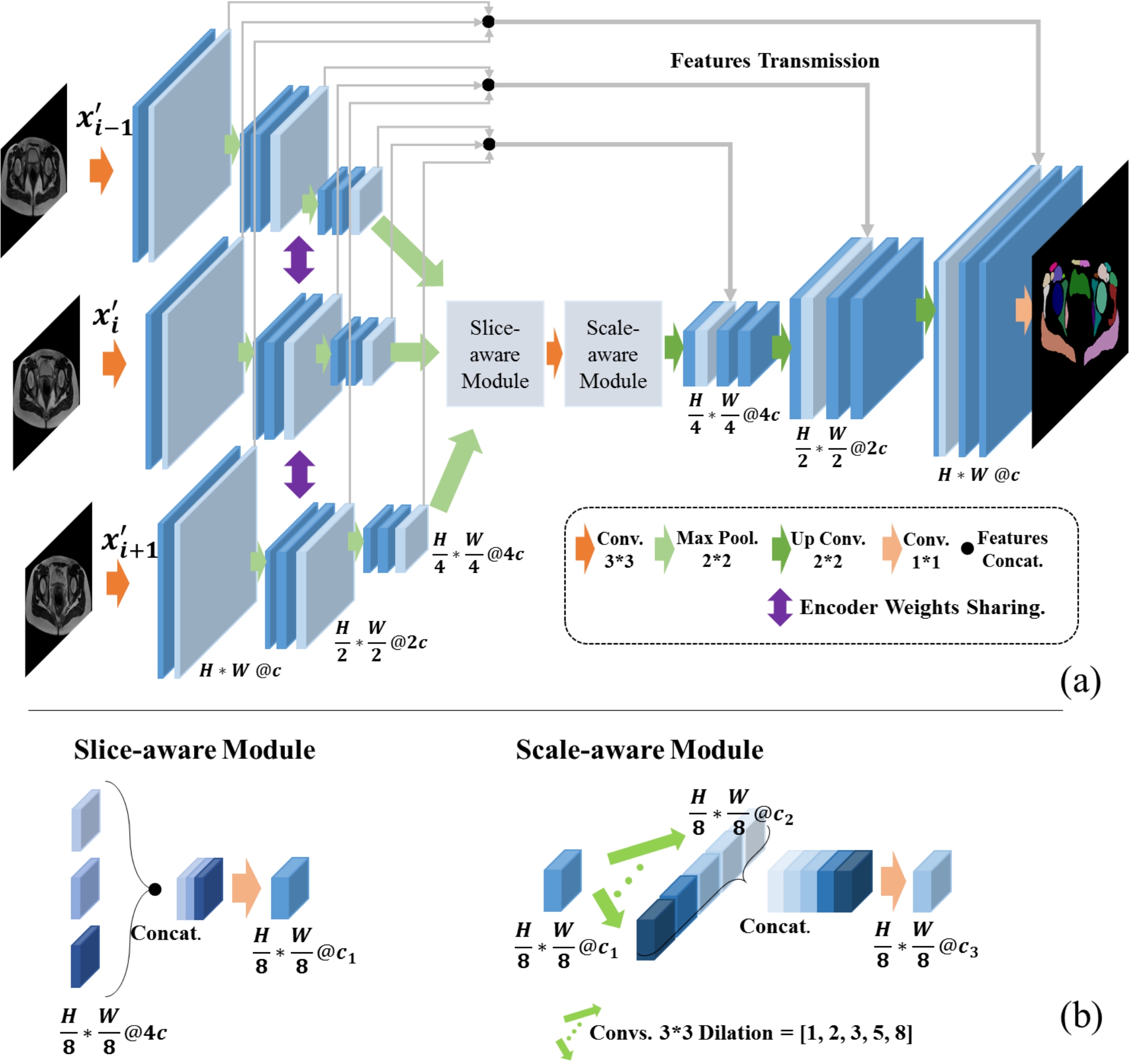
S^2aNet is presented for 3D dense segmentation of 54 organs and musculoskeletal structures in female pelvic MR images. A Scale- aware module is designed to capture the spatial and semantic information of different-scale structures. A Slice-aware module is introduced to model similar spatial relationships of consecutive slices in 3D data. Moreover, S^2aNet leverages a weight-adaptive loss optimization strategy to reinforce the supervision with more discriminative capability on hard samples and categories.
Highlights
-
a weight-adaptive loss optimization strategy is introduced to alleviate difficult samples and the problems of class imbalance.
-
a multislice-aware feature fusion module is proposed to encode and fuse features from different slices by a parameter-sharing mechanism.
-
a parallel multiscale-aware module is designed to extract both spatial information of large-scale categories and semantic information of small-scale categories without losing spatial resolution.
-
To our knowledge, this is the first report to achieve a 3D dense segmentation for pelvic 54 structures on MRI.
Results
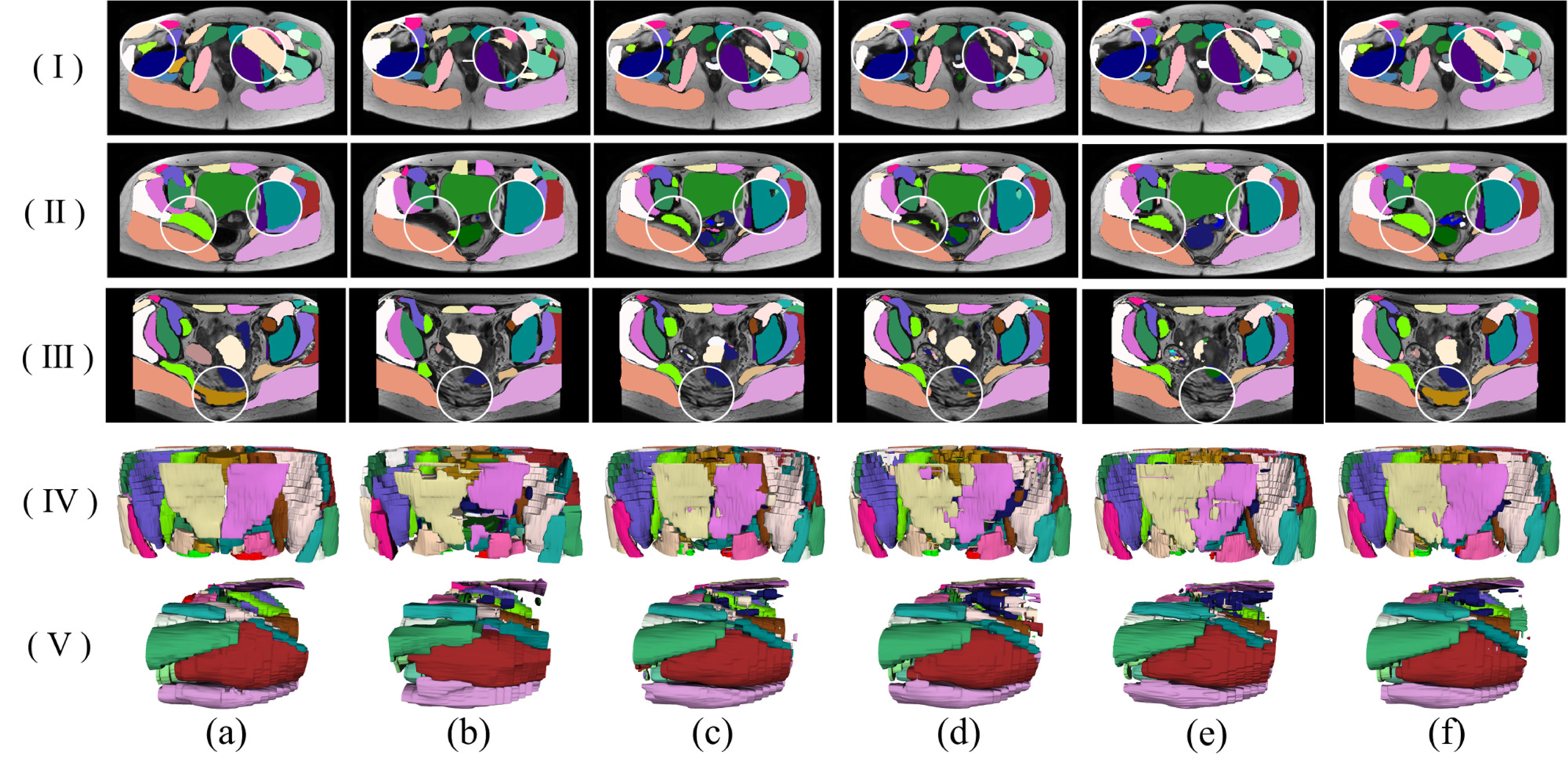
Experiments have been performed on a pelvic MRI cohort of 27 MR images from 27 patient cases. Across the cohort and 54 categories of organs and musculoskeletal structures manually delineated, S^2aNet was shown to outperform the UNet framework and other state-of-the-art fully convolutional networks in terms of sensitivity, Dice similarity coefficient and relative volume difference.
The segmentation results are given below:
Installation and Usage
You need to config the environment firstly, install python and corresponding packages, including torch, opencv, SimpleITK, and so on.
For independent evaluation, run
python A5_test.pyand if you want to generate the predicted results, run
python A6_inference25D.py or if you only want to infer in 2D mode:
python A6_inference2D.py Our network architecture files are Scale_slice_awareNet.py and net_msacunet.py, and both of them are the same.
You can also train the network on your data from scratch.
python A4_trainNet_macunet.py!!! remember to adjust the data path or others privately to yours.
Citation
If the project helps your research, please cite the following paper:
@article{yan2022scale,
title={Scale-and Slice-aware Net (S2aNet) for 3D segmentation of organs and musculoskeletal structures in pelvic MRI},
author={Yan, Chaoyang and Lu, Jing-Jing and Chen, Kang and Wang, Lei and Lu, Haoda and Yu, Li and Sun, Mengyan and Xu, Jun},
journal={Magnetic Resonance in Medicine},
volume={87},
number={1},
pages={431--445},
year={2022},
publisher={Wiley Online Library}
}Acknowledgement
I cherished the memories in AIMLab and thank you all for the wonderful time.
Contact
If you have any problems, just raise an issue in this repo.