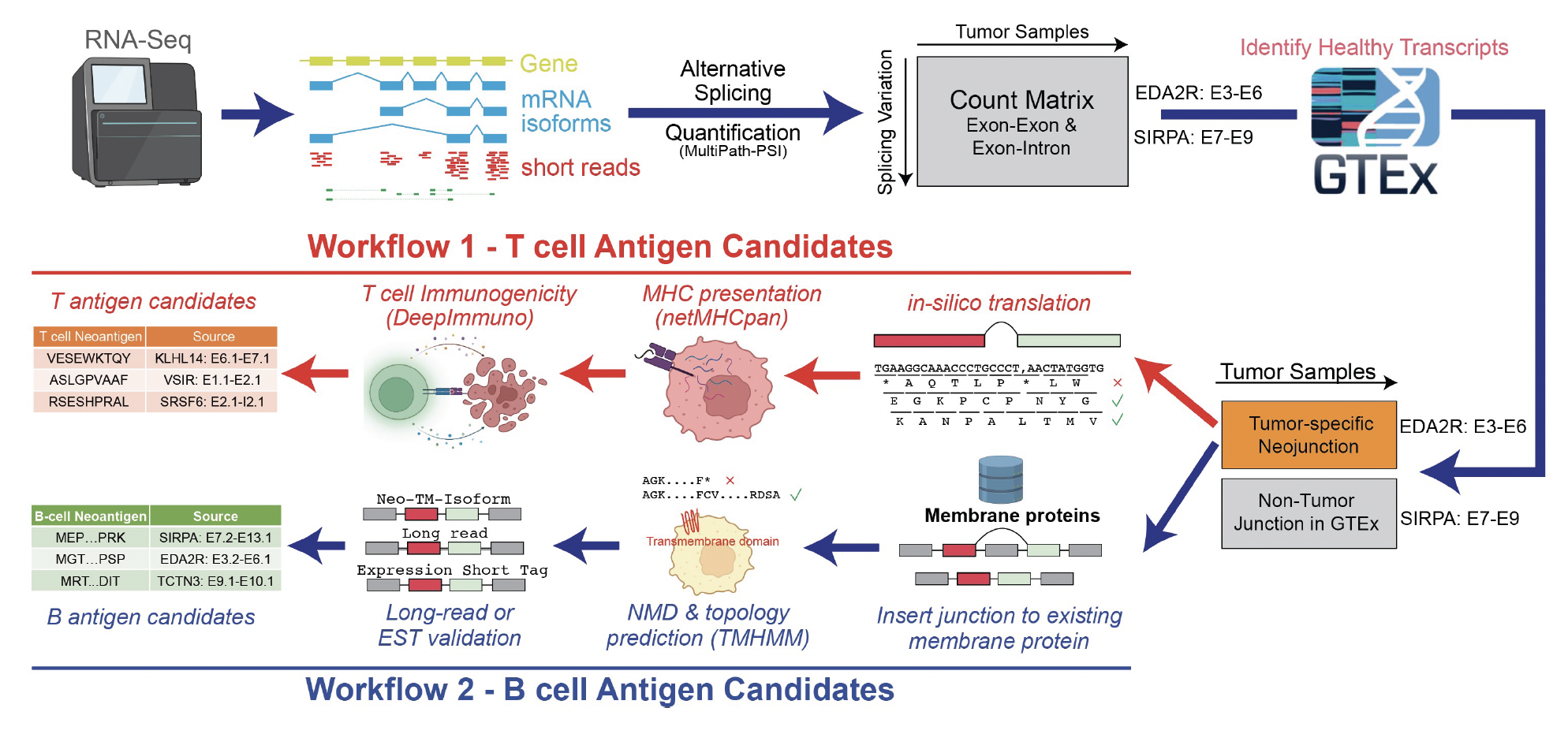
Splicing Neo Antigen Finder (SNAF) is an easy-to-use Python package to identify splicing-derived tumor neoantigens from RNA sequencing data, it can predict, prioritize and visualize MHC-bound neoantigen for T cell (T antigen) and altered surface protein for B cell (B antigen).
See Full Documentation.
Particulary, to avoid hassle, it is advisable to start a fresh conda environment with python 3.7 on a Linux system, and use this specific git commit (I am actively
maintaining it) instead of the stable pypi version.
pip install git+https://github.com/frankligy/SNAF.git@e23ce39512a1a7f58c74e59b4b7cedc89248b908Simply put, user needs to supply a folder with bam files, and the HLA type assciated with each patient (using your favorite HLA typing tool). And it will generate predicted immunogenic MHC-bound peptides and altered surface protein. Moreover, there's a myriad of convenient function that enables users to conduct survival analysis, association analysis and publication-quality visualiztion. Check our tutorials for more detail.
-
RNA-SPRINT: Infering splicing factor activity from RNA-Seq.
-
BayesTS: BayesTS can infer tumor specificity for any features (gene, protein, splicing junction, etc).
-
DeepImmuno: DeepImmuno can infer immunogenicity for peptide-MHC complex.
Guangyuan Li et al. ,Splicing neoantigen discovery with SNAF reveals shared targets for cancer immunotherapy.Sci. Transl. Med.16,eade2886(2024).DOI:10.1126/scitranslmed.ade2886 (https://www.science.org/doi/10.1126/scitranslmed.ade2886)
Guangyuan(Frank) Li
Email: li2g2@mail.uc.edu
PhD student, Biomedical Informatics
Cincinnati Children’s Hospital Medical Center(CCHMC)
University of Cincinnati, College of Medicine