Reconstructing the 3D geometry of the surgical site and detecting instruments within it are important tasks for surgical navigation systems and robotic surgery automation. Traditional approaches treat each problem in isolation and do not account for the intrinsic relationship between segmentation and stereo matching. In this paper, we present a learning-based framework that jointly estimates disparity and binary tool segmentation masks. The core component of our architecture is a shared feature encoder which allows strong interaction between the aforementioned tasks. Experimentally, we train two variants of our network with different capacities and explore different training schemes including both multi-task and single-task learning. Our results show that supervising the segmentation task improves our network’s disparity estimation accuracy. We demonstrate a domain adaptation scheme where we supervise the segmentation task with monocular data and achieve domain adaptation of the adjacent disparity task, reducing disparity End-Point-Error and depth mean absolute error by 77.73% and 61.73% respectively compared to the pre-trained baseline model. Our best overall multi-task model, trained with both disparity and segmentation data in subsequent phases, achieves 89.15% mean Intersection-over-Union in RIS and 3.18 millimetre depth mean absolute error in SCARED test sets. Our proposed multi-task architecture is real-time, able to process (1280x1024) stereo input and simultaneously estimate disparity maps and segmentation masks at 22 frames per second.
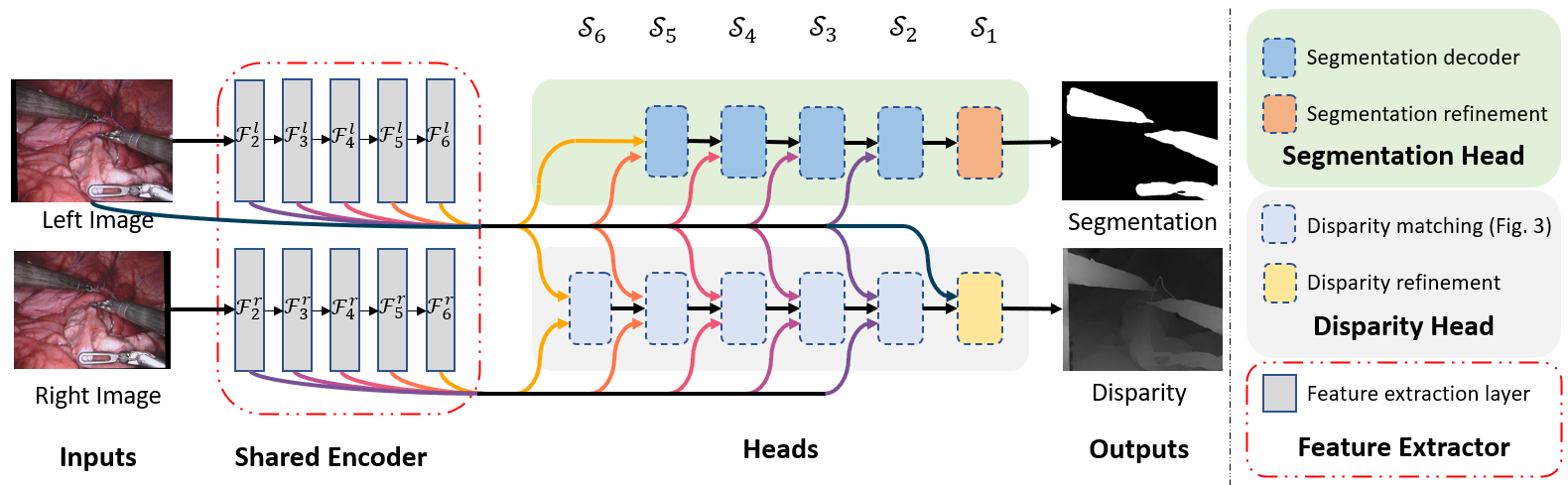 The architecture of our multi-task framework with both heads attached. Left and right rectified images are processed by the shared feature encoder which outputs feature maps Fi at different scales Si. Those features are inputs to each task specific sub-network (head) which make the final predictions. The modular design allows us to interchange or remove part of the network.
The architecture of our multi-task framework with both heads attached. Left and right rectified images are processed by the shared feature encoder which outputs feature maps Fi at different scales Si. Those features are inputs to each task specific sub-network (head) which make the final predictions. The modular design allows us to interchange or remove part of the network.
| SCARED Dataset 8 Keyframe 4 | RIS Dataset d keyframe 264 |
|---|---|
 |
 |
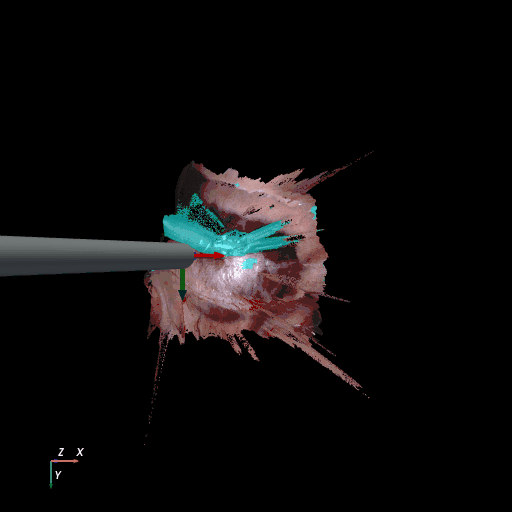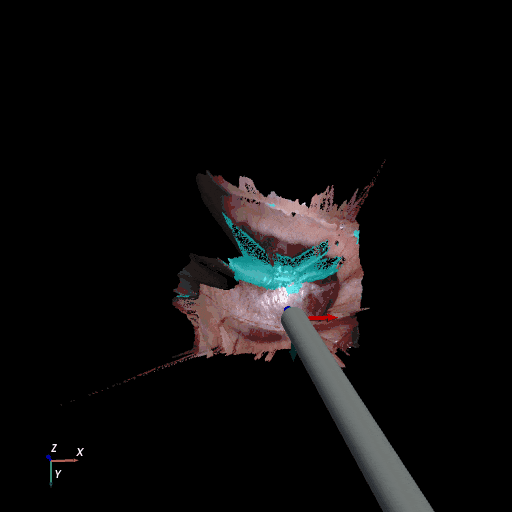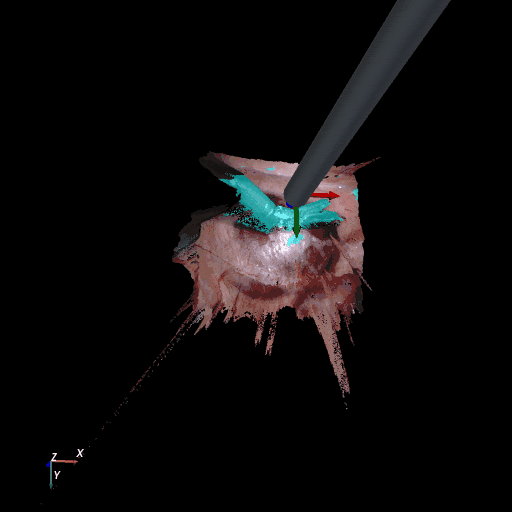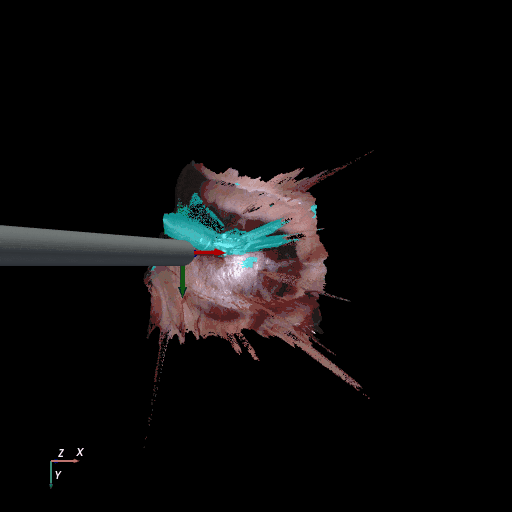
3D reconstruction of SCARED and RIS2017 samples based on the output of our method. Disparity predictions are reconstructed in 3D and binary tool segmentation masks are used to color tool points in shades of cyan. A virtual camera follows a circular trajectory around the endoscope to show details of the final outcome. The model used to estimate both the segmentation and disparity is the light-phase3-multi2seg.
We provide 2 unofficial toolkits to generate disparity samples for SCARED and stereo rectify the RIS2017 dataset which you will need if you want to reproduce that come with this repository. Please visit the repositories bellow:
If you are using this model or any of the provided code for your research, consider citting our work.
@ARTICLE{9791423,
author={Psychogyios, Dimitrios and Mazomenos, Evangelos and Vasconcelos, Francisco and Stoyanov, Danail},
journal={IEEE Transactions on Medical Imaging},
title={MSDESIS: Multi-task stereo disparity estimation and surgical instrument segmentation},
year={2022},
volume={},
number={},
pages={1-1},
doi={10.1109/TMI.2022.3181229}}
- MSDESIS: Multi-task stereo disparity estimation and surgical instrument segmentation
We used Anaconda to handle dependences. Assuming that you have anaconda already installed on your machine, use the following command to create a conda environment suitable to run our code.
conda create -n msdesis --file requirements.txt -c conda-forge pytorch
You can download all the models from here.
unpack the zip and rename, rename the extracted folder to weights and place to
under in the root directory of this repository.
The repository provide a number of inference scripts to facilitate application of our method providing different ways to feed the network with data.
The inference_csv.py script that reads a csv file containing the input stereo
rectified frames as well as the desired location to store the output disparities
and binary tool segmentation masks. You can use the create_io_csv.py script to
create those input .csv files.
To create the input .csv file using the create_io_csv script run the following
command
python -m inference_scripts.create_io_csv left_dir right_dir [--out_path] [--disparity_out_dir] [--segmentation_out_dir]left_dirdirectory where all the left rectified frames are storedright_dirdirectory where all the right rectified frames are stored--out_dirpath to store the output .csv file, default = paths.csv--disparity_out_dirdirectory under which the output disparities predictions will be stored.--segmentation_out_dirdirectory under which the output segmentation predictions will be stored.
--disparity_out_dir and --segmentation_out_dir indicate only the destination
directory under which the output will be stored. The outputs will have the same name
as the left sample.
left_dir and right_dir point to the directory inputs samples are stored. The
script searches for .png files and sorts them. For the script to work properly make
sure your input files are .pngs and for every .png files in left directory their
is a corresponding .png file in the right rectory.
python -m inference_scripts.inference_csv root_input_path root_output_path filelist_path model_type model_weights [--mode] [--cuda_id] [--segmentation_threshold] [--disparity_scale_factor] [--combine] [--jobs] [--use_amp]root_input_pathPath to append add in front of all input paths in csvroot_output_pathPath to append add in front of all output paths in csvfilelist_pathThe path to .csv file containing input and output pathsmodel_typeEncoder type to use, coices=['light', 'resnet34'], default = 'light'model_weightsPath to model weights--modeMode to run the multitask network, choices=['multitask', 'segmentation', 'disparity'], default='multitask'--cuda_idCuda device to use', default=0--segmentation_thresholdSegmentation binary threshold', default=0.5--disparity_scale_factorDisparity scale factor to multiply the original disparity before saving in as uint16 .png, default=128.0--jobsNumber of concurent works used to save output images.--use_ampRun inference in float16. To use this you need an AMP enabled GPU
Good to Know:
- For this script to work you need to make sure that the input samples are rectified.
- The script does not pad the inputs, you need to make sure that the dimensions of the input samples can be divided by 32.
python -m inference_scripts.inference_videos video_path out_path model_type [--max_disparity] [--mode] [--cuda_id] [--segmentation_threshold] [--disparity_scale_factor] [--calib] [--rectification_alpha] [--stacking]This script allows you to run inference directly on a video and export the results to a video making easy qualitative visualization. The script is very versatile and can rectify videos on the fly as long as the stereo calibration parameters are provided.
video_pathPath to input stereo videoout_pathPath to save video showing the network's outputmodel_typeType of encoder to use, choices=['light', 'resnet34'] default='light'model_weightsPath to model weights--max_disparityMaximum disparity search range, default=320. For our 2D network this values should be the same as the one used during training--modeMode to run the multitask network, choices=['multitask', 'segmentation', 'disparity'], default='multitask'--cuda_idCuda device to use', default=0--segmentation_thresholdSegmentation binary threshold, default=0.5--disparity_scale_factorDisparity scale factor to multiply the original disparity, default=1, because we save the output as a video and not as png, this is only useful when the disparity range is over 255--calibPath to stereo calibration .json file, to stereo rectify the input video. If this is not set, the input video is considered rectified and its frames are fed directly to the network--rectification_alphaStereo rectification alpha to use in the stereo rectification, default=-1.0--stackingSpecify if left and right stereo frames are stacked horizontally or vertically in the input video, choices=['horizontal', 'vertical'], default='horizontal'--save_imagesWherether to save images instead of video, default=False
The following scripts can be used to evaluate the performance of the network in RIS2017 and SCARED datasets. None of those scripts are from the original authors of those datasets. The scripts output a summary of performance, similar to what is reported in the target dataset. Both scripts use the network to process input frames and then compare the outputs with the reference samples from the dataset.
For the following script to work you need to have a copy of RIS dataset and configure
the datasets/dataset_paths.json file.
python -m evaluation_scripts.evaluate_ris model_type model_weights --save_predictions --csv --cuda_id --segmentation_thresholdmodel_typeEncoder type to use, choices=['light', 'resnet34'], default='light'model_weightsPath to model weights--save_predictionsDirectory to save network output, if not specified, the script will only save the score.--csvPath to save evalution scores, default='./ris17_scores.csv'--cuda_idCuda device to use, default=0--segmentation_thresholdSegmentation binary threshold, default=0.5
For the following script to work you need to have a rectified copy of SCARED dataset
with disparity and configure the datasets/dataset_paths.json file. To generate
disparity such dataset have a loot at scared-toolkit
The script can evaluate both disparity and optionally depth-maps. However results
computed from this script are not compatible with the SCARED evalutation
protocol and do not correspond to the figures in our paper. To recreate results
from the paper you need to produce disparity images using the inference_csv script
convert those to original depth and compute the error against the provided ground truth
using the scared-toolkit. The scared-toolkit repository provides code to convert
disparity back to original frame of reference and evaluation code that rejects
low coverage frames.
To evaluate loosly on SCARED:
python -m evaluation_scripts.evaluate_scared model_type model_weights --save_predictions --csv --cuda_id --segmentation_threshold --disparity_scale_factor --depthmodel_typeEncoder type to use, choices=['light', 'resnet34'], default='light'model_weightsPath to model weights--save_predictionsDirectory to save network output, if not specified, the script will only save the score.--csvPath to save evalution scores, default='./scared_scores.csv'--cuda_idCuda device to use, default=0--segmentation_thresholdSegmentation binary threshold, default=0.5--disparity_scale_factorDisparity scale factor to multiply the original disparity before saving it as 16bit .png, default=128--depthCalculate depth mean absolute error
There are three main training scripts for Disparity, Segmentation and Multitask training.
We use json files to configure hyperparameter and pass those as arguments when running the scripts.
To recreate our results you can use the configuration scripts provided under the /training_configs/ directory.
- Phase 1: You will need a copy of the FlyingThings3D subset available here. To recreate results you only have to download the rgb images and the disparity ground truth files. You will also need to modify the
flyingthings3dentry of thedatasets/dataset_paths.jsonfile to point to the root folder of your FlyingThings3D dataset. - phase 2-3: To train the network in the scared dataset you will need to download scared and create a keyframe only subset. Instructions and tools on how to do that can be found here. It is important to note that during training we did not use the provided ground truth files because those occasionally have extreme outliers. To recreate the paper's results consider manually cleaning the provided pointcloud of obvious outliers before generating the disparities. In addition when generating disparities you need to use a scale factor of 128. As was the case with Flyingthings3D, you will need to modify the
scaredentry of thedatasets/dataset_paths.jsonfile to point to the root folder to the manipulated SCARED dataset. Additionally you need to train the previous phase model or download their corresponding weights.
The folder structure of the SCARED keyframe data should look like this:
keyframe_scared //root directory, scared needs to point to.
├── disparity //disparity directory, disparities are stored using a scale factor of 128
│ ├── 1_1.png //{dataset_number}_{keyframe_number}.png
│ ├── :
│ └── 7_5.png
├── left_rectified // left rectified image directory
│ ├── 1_1.png
│ ├── :
│ └── 7_5.png
└── right_rectified //right rectified image directory
├── 1_1.png
├── :
└── 7_5.png
python -m training_scripts.train_disparity --default_config --cuda_id--default_config .json file containing hyperparameter configuration
--cuda_id gpu number you want to use, default=0
To recreate the paper's results you will need to have a ph1 disparity model to train the multitask model, or download the pretrained models.
Additionally you will need to have a stereo rectified version of the RIS 2017 dataset. Tools and instructions on how to get one can be found here.
After you have a stereo rectified copy of the RIS, you will then need to modify the ris17_124 entry of the datasets/dataset_paths.json file to point to the train subset of the the stereo rectified RIS.
This should follow the file structured shown bellow:
stereo_rectified_ris_train //root directory, ris17_124 needs to point to.
├── instrument_dataset_1
│ ├── ground_truth
│ │ └── binary
│ │ ├── frame000.png //frame{frame_number}.png
│ │ ├── :
│ │ └── frame224.png
│ ├── left_frame
│ │ ├── frame000.png
│ │ ├── :
│ │ └── frame224.png
│ └── right_frame
│ ├── frame000.png
│ ├── :
│ └── frame224.png
├── instrument_dataset_2
├── instrument_dataset_3
├── instrument_dataset_4
├── instrument_dataset_5
├── instrument_dataset_6
├── instrument_dataset_7
└── instrument_dataset_8
python -m training_scripts.train_multitask --default_config --cuda_id--default_config .json file containing hyperparameter configuration
--cuda_id gpu number you want to use, default=0
Good things to know:
- To train with mixed precision you need to remove the type assertions in kornia's loss functions.
To recreate the paper's results, you will need to have a ph1 disparity model to train the ph2 segmentation model and a ph2 multitask model to train the ph3 segmentation model.
Again for the training scripts to work you need to have a copy of RIS. In our experiments we de-interlaced, cropped and resized the provided samples. Tools to do that are provided here. Additionally you will need to modify the ris17_train_monocular entry of the datasets/dataset_paths.json file to point to the train subset of the training RIS set. The file structure of the monocular ris should be the same as the stereo rectified one.
python -m training_scripts.train_segmentation --default_config --cuda_id--default_config .json file containing hyperparameter configuration
--cuda_id gpu number you want to use, default=0