Detect somatic copy number variants (CNV) in tumour-normal samples using the facets package
- Purpose
- Quick start
- Requirements and Installation
- Input
- Output
- Usage guidelines
- Time and memory footprint
- Citation & Getting help
cnv_facets detects somatic copy number variants (CNVs), i.e., variants private to a tumour sample given a matched or unmatched normal sample. cnv_facets uses next generation sequencing data from whole genome (WGS), whole exome (WEX) and targeted (panel) sequencing experiments. In addition, it estimates tumour purity and ploidy.
The core of cnv_facets is the facets package by R Shen and VE Seshan FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing, Nucleic Acids Res, 2016
The advantage of cnv_facets over the original facets package is the convenience of executing all the necessary steps, from BAM input to VCF output, in a single command line call.
Install with bioconda
conda install cnv_facets
Detect CNVs:
cnv_facets.R -t <tumour.bam> -n <normal.bam> -vcf <snps.vcf.gz> -o <output_prefix>
Get help:
cnv_facets.R -h
cnv_facets runs on the Linux operating system. Windows is not supported
and MacOS could work but some tweaks are necessary.
Installation via the conda package manager is the
recommended route. Options -c bioconda -c conda-forge can be omitted if
bioconda and conda-forge are already registered channels (see below).
It is generally not recommended to install packages in the conda base environment. Better to
install in a dedicated envirnment. E.g.:
conda create -n my_project
conda activate my_project
conda install -c bioconda -c conda-forge cnv_facets
If the above fails with conda: command not found or similar, install conda first.
Follow the official
documentation but
basically, these commands should suite most users:
# See https://conda.io/miniconda.html
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
# Run and follow the prompt on screen
bash Miniconda3-latest-Linux-x86_64.sh
# Add some useful package repositories
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
cnv_facets requires a reasonably recent version of R on a Linux operating system. At the time of this writing, it has been developed and deployed on R 3.5 on CentOS 7.
To compile and install execute:
bash setup.sh --bin_dir </dir/on/path>
Where /dir/on/path is a directory on your PATH where you have permission to
write, e.g., ~/bin.
Required input files:
-
A bam file of the tumour sample
-
A bam file of the normal sample (typically, a blood sample from the same patient)
-
A VCF file of common, polymorphic SNPs. For human samples, a good source is the dbSNP file common_all.vcf.gz. See also NCBI human variation sets in VCF Format.
BAM and VCF files must be sorted and indexed.
USAGE
cnv_facets.R -t <tumour.bam> -n <normal.bam> -vcf <snps.vcf.gz> -o <output_prefix> [...]
This pileup file is generated by cnv_facets.R when run with bam input as in
option 1. If you need to explore different parameter values for CNV detection,
using a pre-made pileup file can save considerable computing time.
Internally, cnv_facets.R uses snp-pileup, a program installed together
with the cnv_facets package.
The pileup is a comma separated file of read counts for the reference and alternate allele at polymorphic SNPs. This file must have the following columns (order of columns is not important, additional columns are ignored):
-
Chromosome Chromosome of the SNP
-
Position Position of the SNP
-
File1R Read depth supporting the REF allele in normal sample
-
File1A Read depth supporting the ALT allele in normal sample
-
File2R Read depth supporting the REF allele in tumour sample
-
File2A Read depth supporting the ALT allele in tumour sample
These are the first lines of the test file test/data/stomach.csv.gz
accompanying the original facets package:
"Chromosome","Position","Ref","Alt","File1R","File1A","File1E","File1D","File2R","File2A","File2E","File2D"
1,69424,N,N,170,117,0,0,158,103,0,0
1,69515,N,N,0,76,0,0,0,77,0,0
1,69536,N,N,103,0,0,0,99,0,0,0
1,808866,N,N,96,0,0,0,133,0,0,0
1,809120,N,N,66,0,0,0,105,0,0,0
USAGE
cnv_facets.R -p <pileup.csv.gz> -o <output_prefix> [...]
The option --out/-o <prefix> determines the name and location of the output
files. For more information refer to the documentation of the
facets package.
<prefix>.vcf.gz
VCF file compressed and indexed of copy number variants. The INFO tags below annotate each variant:
| Tag | Type | Description |
|---|---|---|
| SVTYPE | String | Type of structural variant |
| SVLEN | Integer | Difference in length between REF and ALT alleles |
| END | Integer | End position of the variant described in this record |
| NUM_MARK | Integer | Number of SNPs in the segment |
| NHET | Integer | Number of SNPs that are deemed heterozygous |
| CNLR_MEDIAN | Float | Median log-ratio (logR) of the segment. logR is defined by the log-ratio of total read depth in the tumor versus that in the normal |
| CNLR_MEDIAN_CLUST | Float | Median log-ratio (logR) of the segment cluster. logR is defined by the log-ratio of total read depth in the tumor versus that in the normal |
| MAF_R | Float | Log-odds-ratio (logOR) summary for the segment. logOR is defined by the log-odds ratio of the variant allele count in the tumor versus in the normal |
| MAF_R_CLUST | Float | Log-odds-ratio (logOR) summary for the segment cluster. logOR is defined by the log-odds ratio of the variant allele count in the tumor versus that in the normal |
| SEGCLUST | Integer | Segment cluster to which the segment belongs |
| CF_EM | Float | Cellular fraction, fraction of DNA associated with the aberrant genotype. Set to 1 for normal diploid. See also issue #17 |
| TCN_EM | Integer | Total copy number. 2 for normal diploid |
| LCN_EM | Integer | Lesser (minor) copy number. 1 for normal diploid |
| CNV_ANN | String | Annotation features assigned to this CNV |
The header of the VCF file also stores the estimates of tumour purity and ploidy and the average insert size of the normal library if using paired-end BAM input.
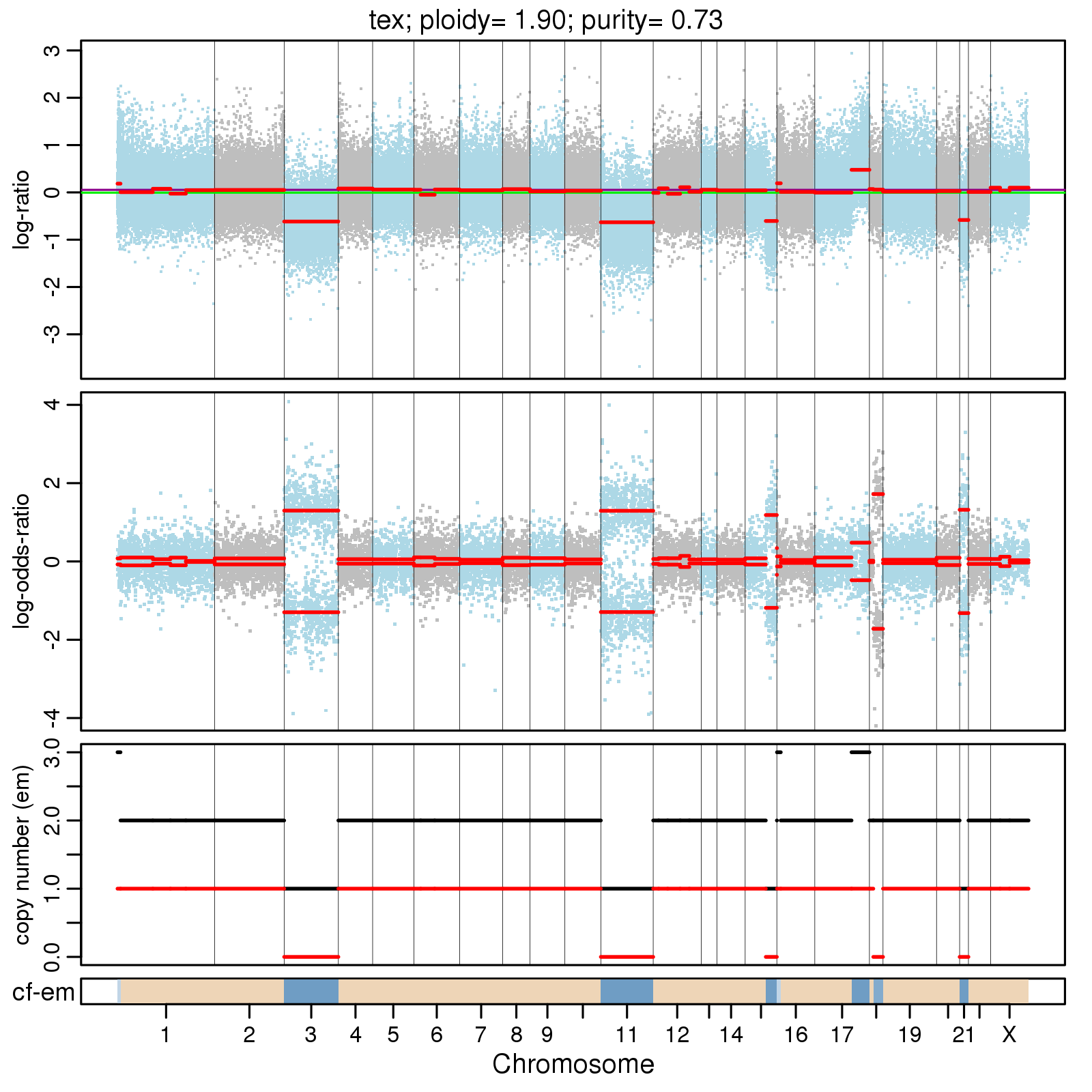
<prefix>.cnv.png
Summary plot of CNVs across the genome, for example:
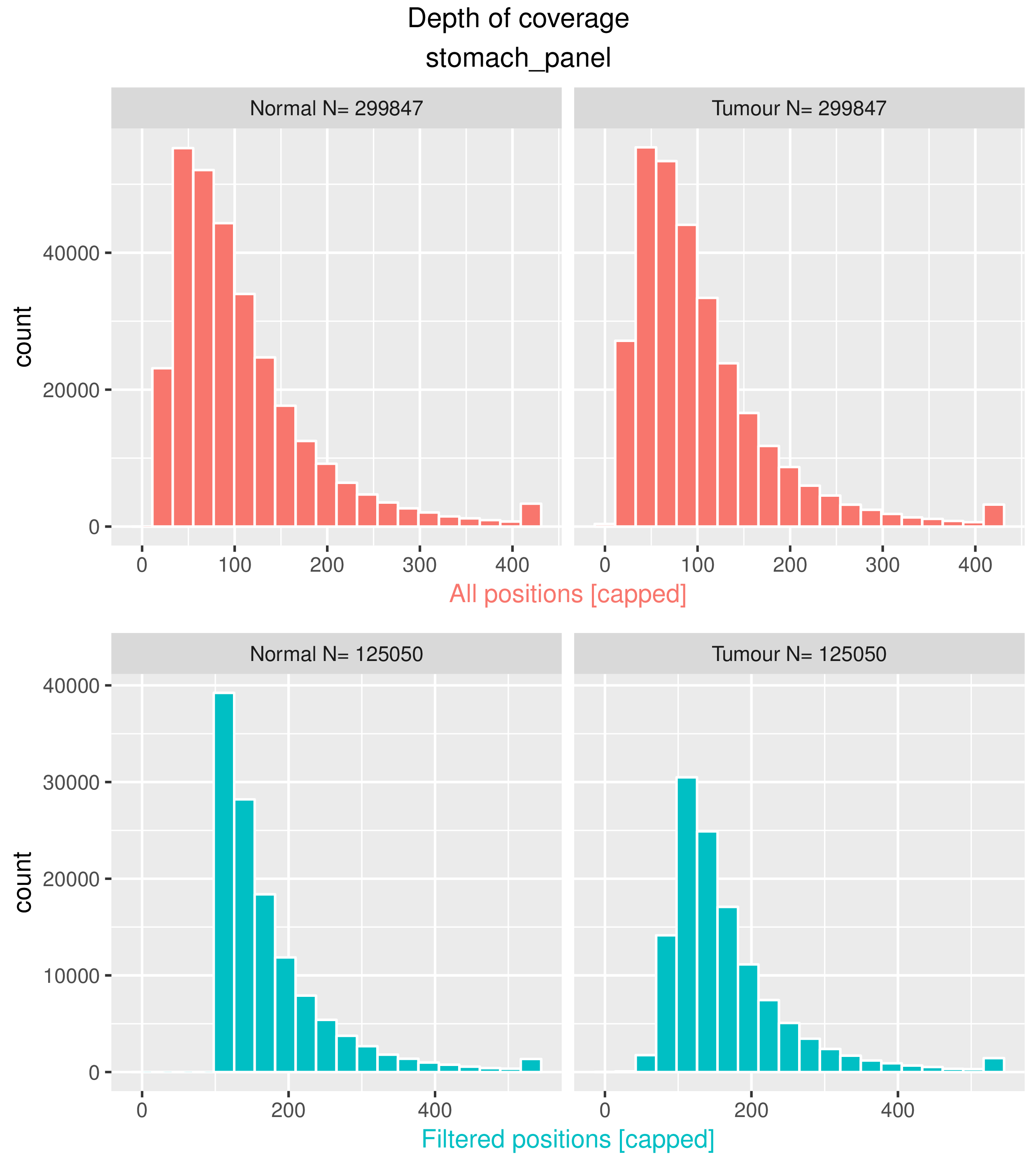
<prefix>.cov.pdf
Histograms of the distribution of read depth (coverage) across all the position in the tumour and normal sample, before and after filtering positions. These plots are useful to assess whether the sequencing depth and depth of covarage thresholds are appropriate.
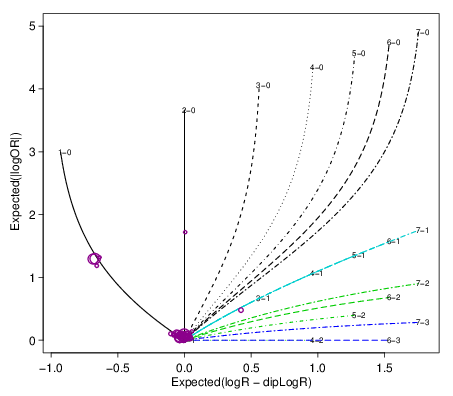
<prefix>.spider.pdf
This is a diagnostic plot to check how well the copy number fits
work The estimated segment summaries are plotted as circles
where the size of the circle increases with the number of loci in
the segment. The expected value for various integer copy number
states are drawn as curves for purity ranging from 0 to 0.95. For
a good fit, the segment summaries should be close to one of the
lines. (Description from facets::logRlogORspider). For example:
<prefix>.csv.gz
File of nucleotide counts at each SNP in normal and tumour sample.
--depth
Use the histograms of depth to set appropriate thresholds. Consider also the option
--targets for targeted sequence libraries.
--cval
Critical values for segmentation in pre-processing and processing. Larger values reduce segmentation. [25 150] is facets default based on exome data. For whole genome consider increasing to [25 400] and for targeted sequencing consider reducing them. Default 25 150
--nbhd-snp
If an interval of size nbhd-snp contains more than one SNP, sample a random one. This sampling reduces the SNP serial correlation. This value should be similar to the median insert size of the libraries. 250 is facets default based on exome data. For whole genome consider increasing to 500 and for target sequencing decrease to 150. Default 250
- CNLR_MEDIAN_CLUST
USe this VCF tag to filter for records where the difference in read depth
coverage between tumour and normal. The tag CNLR_MEDIAN should be well
correlated with CNLR_MEDIAN_CLUST so using one or the other should not make
much difference. Use the plot of CNV profile, log-ratio panel of
<prefix>.cnv.png to decide on a sensible thresholds.
- MAF_R_CLUST
Use this VCF tag to filter for CNVs significant difference in tumour allele
frequency. Use the plot of CNV profile, log-odds-ratio panel of <prefix>.cnv.png
to decide on a sensible thresholds. As above MAF_R_CLUST is correlated with MAF_R.
The analysis of a whole genome sequence where the
tumour is sequenced at ~80x (~2 billion reads, BAM file ~200 GB) and the normal
at ~40x (~1 billion reads, BAM files ~100 GB) with ~37 million SNPs (from dbSNP
common_all_20180418.vcf.gz) and with no filtering on read depth and read
quality requires:
-
5 hours to prepare the SNP pileup with small memory footprint. Time is mostly driven by the size of the BAM files. To speed-up the pileup consider the option
--snp-nprocsto parallelize across chromosomes. -
1 hour and ~15 GB of memory for the actual detection of CNVs starting from the pileup. Time and memory is mostly driven by the number of SNPs
If using cnv_facets please cite
-
the URL of this repository and
-
The publication of the facets package FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing, Nucleic Acids Res, 2016
Any and all comment and questions can be sent to one or more of the following recipients:
-
Open an issue at github.com/dariober/cnv_facets)
-
For questions specific to the FACETS package and CNV calling open an issue at https://github.com/ddmskcc/facets
-
Post a question at https://www.biostars.org/ (you may want to notify me by sending an email to
dario <dot> beraldi <at> gmail <dot> com)