These files are associated with the following publication:
DT Yang, AM Gronenborn, LT Chong, “Integrating Fluorinated, Aromatic Amino Acids into the Framework of the AMBER ff15ipq Force Field.” J. Phys. Chem. A, 2022, in press; https://doi.org/10.1021/acs.jpca.2c00255
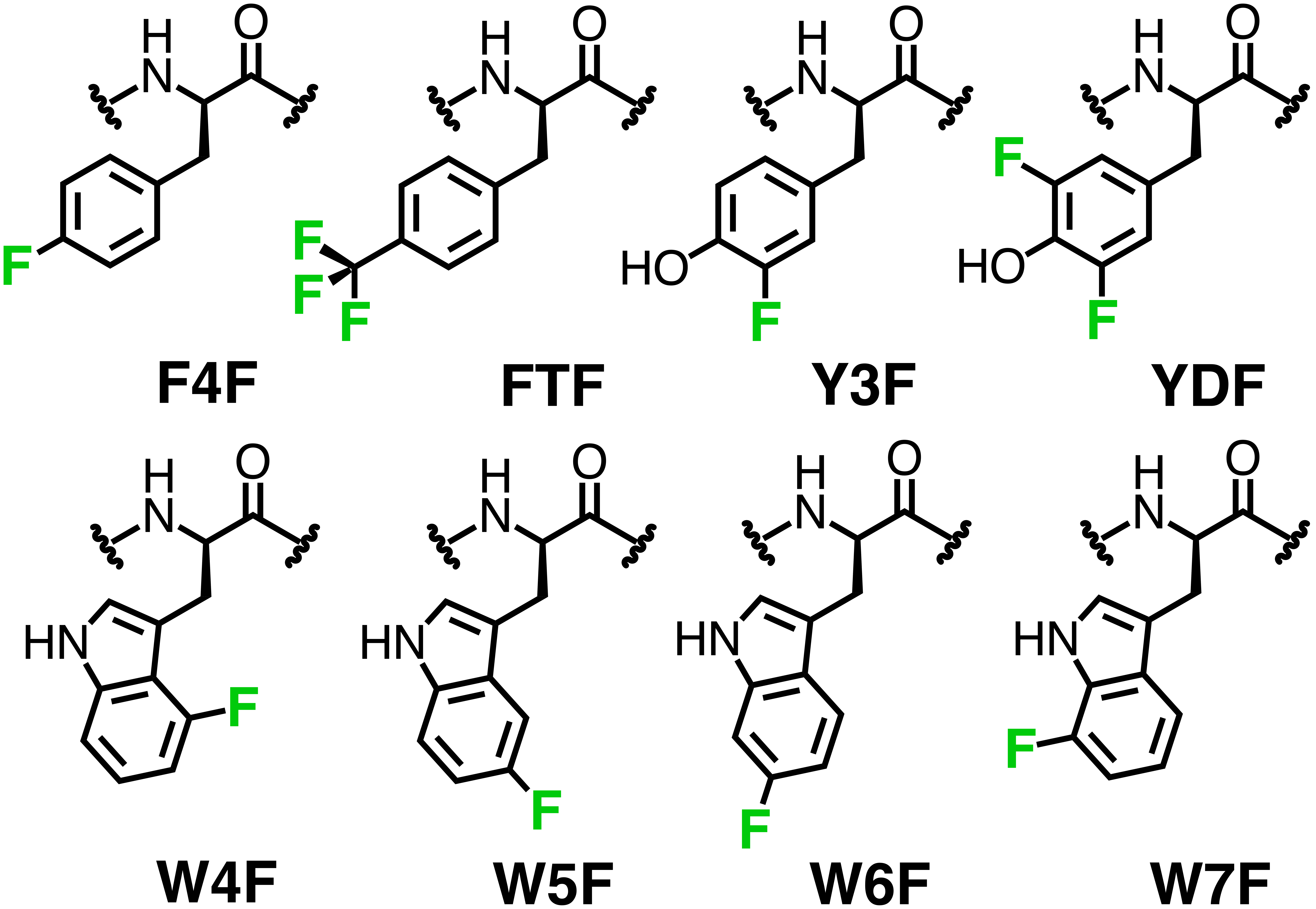
- force field parameters for fluorinated amino acids commonly used with 19F NMR experiments
- see final_parameters/ for the AMBER compatible frcmod (bonded parameters) and leap (ipq charges) files for use with the ff15ipq protein force field, along with instructions for how to use these files
- not all original files were included due to size limitations
- raw trajectory data is available upon reasonable request: please email dty7@pitt.edu
- see analysis/ for Python scripts used for analysis and figure generation
- for fluorine relaxation calculations, see https://github.com/darianyang/fluorelax