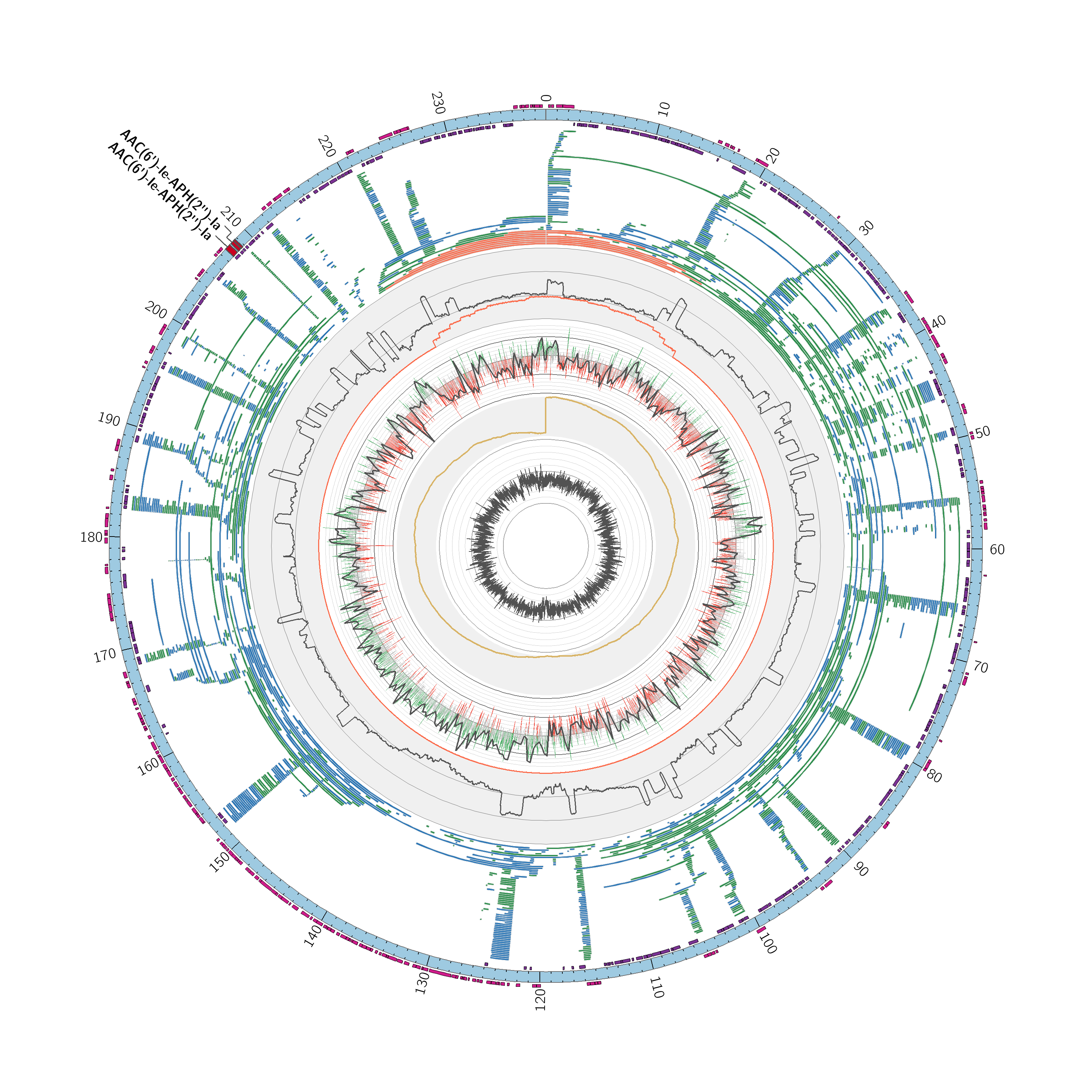
This pipeline idenfitifes circular plasmids in in bacterial genome assemblies using long reads.
It includes the following steps
- Gene prediction with Glimmer3
- Identification of antibiotic resistance genes in the CARD Database RGI
- Long read alignment against assembly
- Coverage analysis with Mosdepth
- GC Content and GC Skew
- Identification of reads that overlap the gap in the plasmid, indicating circular reads
It is created with nextflow, an application to create complex pipelines with repository integration
- Linux or Mac OS (Not tested on Windows, might work with docker)
- Java 8.x
- Install nextflow
curl -s https://get.nextflow.io | bash
This creates the nextflow executable in the current directory
- Download pipeline
You can either get the latest version by cloning this repository
git clone https://github.com/caspargross/plasmident
or download on of the releases.
- Download dependencies
All the dependencies for this pipeline can be downloaded in a docker container.
docker pull caspargross/plasmident
Alternative dependency installations:
The pipeline requires an input file with a sample id (string) and paths for the assembly file in .fasta format and long reads in .fastq or .fastq.gz. The paths can either be absolute or relative to the launch directory. In normal configuration (with docker), it is not possible to follow symbolic links.
The file must be tab-separated and have the following format
| id | assembly | lr |
|---|---|---|
| myid1 | /path/to/assembly1.fasta | /path/to/reads1.fastq.gz |
| myid2 | /path/to/assembly2.fasta | /path/to/reads2.fastq.gz |
The pipeline is started with the following command:
nextflow run plasmident --input read_locations.tsv
There are other run profiles for specific environments.
--outDirPath of output folder--seqPaddingNumber of bases added at contig edges to improve long read alignment [Default: 1000]--covWindowMoving window size for coverage and gc content calculation [Default: 50]--cpuNumber of threads used per process--targetCovLarge read files are subsampled to this target coverage to speed up the process [Default: 50]