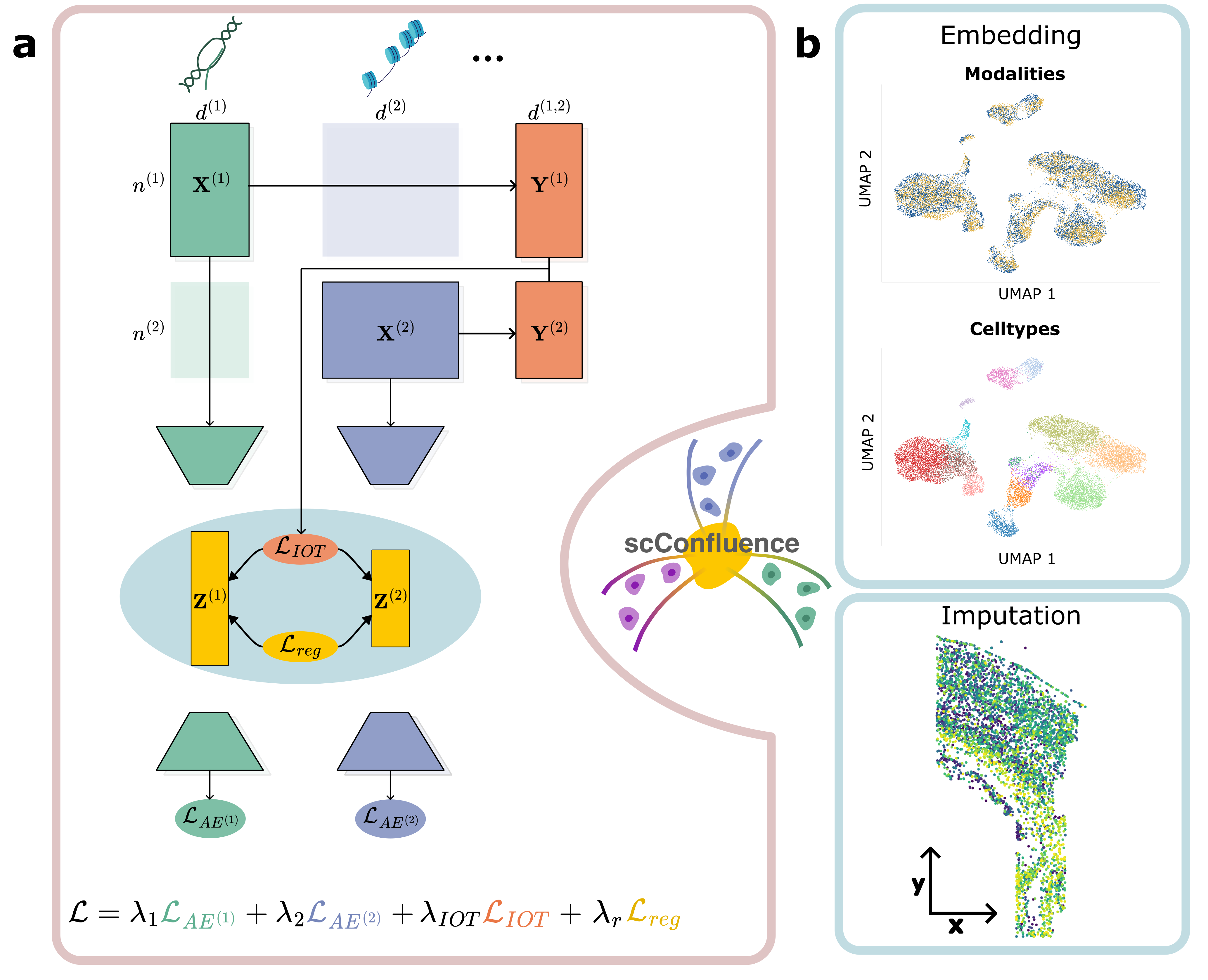
scConfluence is a novel method for the integration of unpaired multiomics data combining uncoupled autoencoders and Inverse Optimal Transport to learn low-dimensional cell embeddings. These embeddings can then be used for visualization and clustering, useful for discovering subpopulations of cells, and for imputation of features across modalities. Read the preprint!
scConfluence is implemented as a Python package seamlessly integrated within the scverse ecosystem, in particular Muon and Scanpy.
On all operating systems, the easiest way to install scConfluence is via PyPI. Installation should typically take a minute and is continuously tested with Python 3.10 on an Ubuntu virtual machine.
pip install scconfluencegit clone git@github.com:cantinilab/scconfluence.git
cd scconfluence
pip install .pip install pytest
pytest .scConfluence takes as an input a MuData object and populates its obsm field
with the latent embeddings. Visit http://scconfluence.rtfd.io/ for
more documentation and tutorials (which can be found now in the tutorials folder of
this repository).
You may download a preprocessed 10X Multiome demo dataset here.
A GPU is not required for the method to run, but is strongly recommended.
import scconfluence
import mudata as md
import scanpy as sc
from scipy.spatial.distance import cdist
# Load data into a Muon object.
mdata = md.read_h5mu("my_data.h5mu")
# Compute the cross-modality distance matrix using connected features
mdata.uns["cross_modality1+modality2"] = cdist(mdata["modality1"].obsm["cm_features"],
mdata["modality2"].obsm["cm_features"])
mdata.uns["cross_keys"] = ["cross_modality1+modality2"]
# Initialize and train the model.
autoencoders = {"modality1": scconfluence.model.AutoEncoder(mdata["modality1"],
modality="modality1"),
"modality2": scconfluence.model.AutoEncoder(mdata["modality2"],
modality="modality2")}
model = scconfluence.model.ScConfluence(mdata, unimodal_aes=autoencoders)
model.fit(save_path="results")
mdata.obsm["latent"] = model.get_latent().loc[mdata.obs_names]
# Visualize the embedding with UMAP.
sc.pp.neighbors(mdata, use_rep="latent")
sc.tl.umap(mdata)
sc.pl.umap(mdata)https://www.biorxiv.org/content/10.1101/2024.02.26.582051v1
@article {Samaran2024unpaired,
author = {Jules Samaran and Gabriel Peyre and Laura Cantini},
title = {scConfluence : single-cell diagonal integration with regularized Inverse Optimal Transport on weakly connected features},
year = {2024},
doi = {10.1101/2024.02.26.582051},
publisher = {Cold Spring Harbor Laboratory},
journal = {bioRxiv}
}If you're looking for the repository with code to reproduce the experiments in our preprint, here it is!