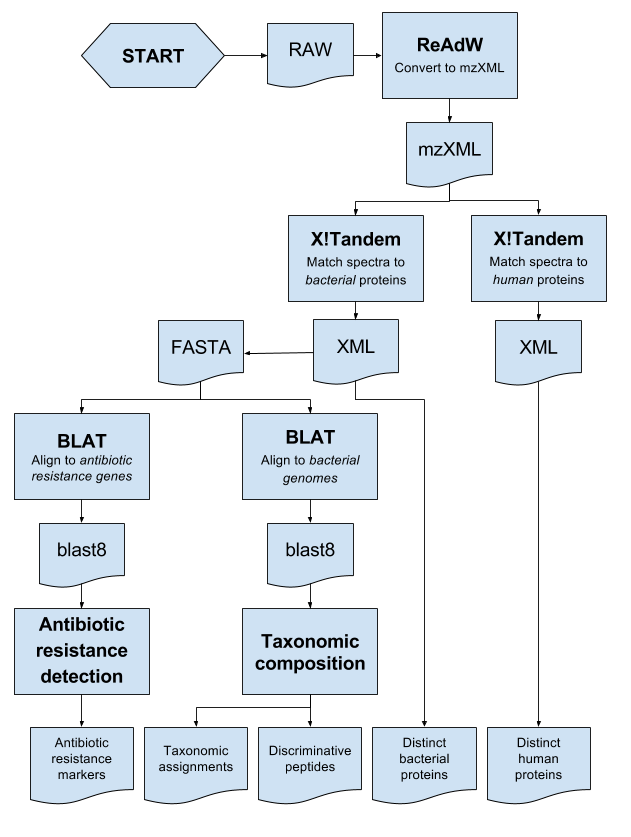
This repository contains the components of "The Tailored Treatment Proteotyping Pipeline", TPARTY, for automated proteotyping analysis of samples in The Tailored Treatment project.
For details on the implementation, please refer to the somewhat self-documenting Snakemake script, located in the Snakemake folder of the repository.
- Authors
Fredrik Boulund
- Contact
- License
ISC
This is the README file for the TTT Proteotyping Pipeline, called TPARTY. Refer to the online documentation for instructions on how to use it. The code for the project is published as open-source under the ISC license and you are welcome to look at, suggest improvements, or download and improve/contribute to the code via the project's Bitbucket page.
Detailed installation instructions are available in the online documentation.
Clone the repository to obtain a copy of the code and scripts:
$ hg clone https://bitbucket.org/chalmersmathbioinformatics/tpartyThe pipeline depends on a number of Python packages and external programs, most notably:
- ReAdW (raw to mzXML conversion)
- X!Tandem mass-spectrometry search engine (mzXML to peptides)
- BLAT (sequence alignment)
- TCUP
If you find the TTT Proteotyping Pipeline useful, please cite us! The method is described in the following papers:
TCUP: Typing and characterization of bacteria using bottom-up tandem mass spectrometry proteomics
Mol Cell Proteomics mcp.M116.061721. First Published on April 18, 2017,
DOI: http://dx.doi.org/10.1074/mcp.M116.061721
Keywords: mass spectrometry, proteomics, microbial identification, pathogenic bacteria, antibiotic resistance detection, LC-MS/MS
and
Proteotyping: Proteomic characterization, classification and identification of microorganisms – A prospectus
Systematic and Applied Microbiology, Volume 38, Issue 4, June 2015, Pages 246-257, ISSN 0723-2020
DOI: http://dx.doi.org/10.1016/j.syapm.2015.03.006
http://www.sciencedirect.com/science/article/pii/S0723202015000491
Keywords: Proteotyping; Proteomics; Mass spectrometry; Microbial systematics