Deep learning is an important machine learning technique that helps computers identify objects in images. Medical images are used in diagnosis and treatment of diseases, injuries, and congenital abnormalities.
Deep learning can be used to detect diseases like cancer by finding tumor cells through medical images. Deep learning also has the potential to improve the quality of medical care by segmenting organs during surgery or scanning patients for signs of cancer or other ailments.
The goal of this repository is to create a deep learning model to segment a liver from a public CT scan dataset. After completing this series, you will be able to create the same model that segments the liver, as well as use the same principle to segment other organs or tumors from CT scans or MRIs.
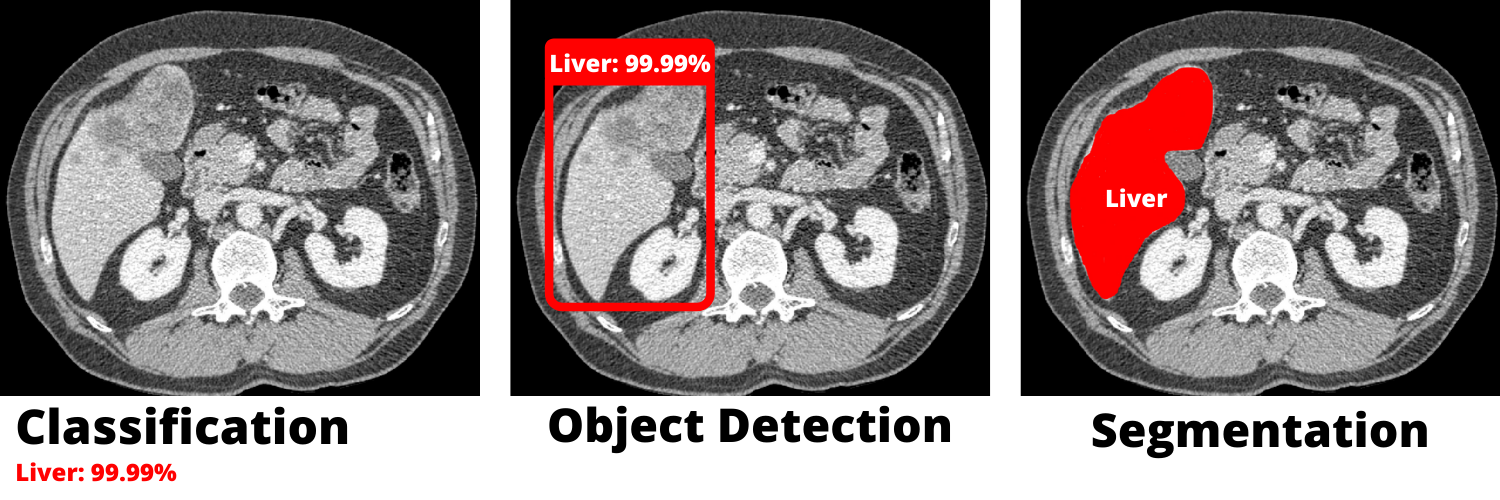
Segmentation is one of various applications in computer vision. It does not only figure out the nature of the object (classifying) or draws a box around it (object detection), instead it adds a whole layer above the original DICOM file. both images has the same dimensions and the latter emphasizes the object we are working on.
The picture below shows the difference between the techniques mentioned above.
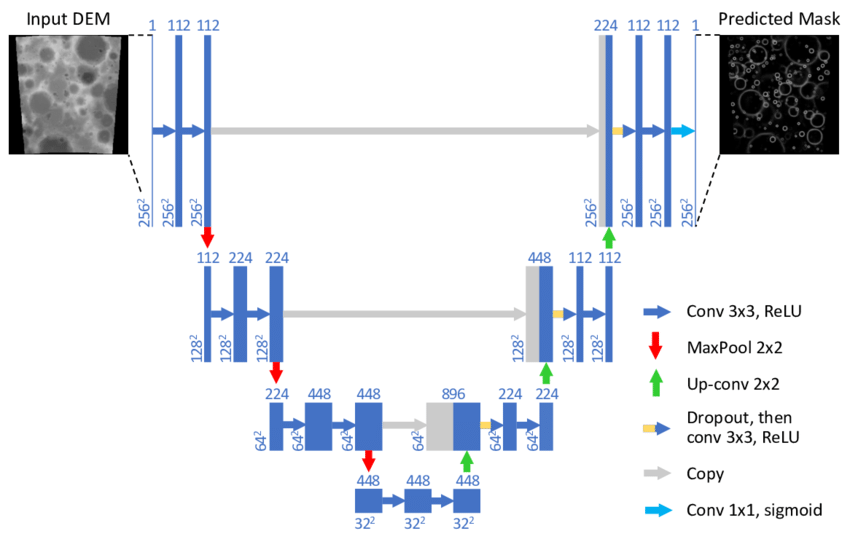
The u-net is Convolutional Networks for Biomedical Image Segmentation. It is a convolutional neural network architecture that expanded with few changes in the CNN architecture. it was initially conceived for medical. It was invented to deal with biomedical images where the target is not only to classify whether there is an infection or not but also to identify the area of infection.. the U-Net has later been adopted in multiple fields thanks to it's effeciency.
The illustration below shows the architecture of the U-Net
Using frameworks is fundamental. using the right frameworks for the job could immensely simplify the job.
For this project we used the following technologies.
-
MONAI: Medical Open Networkfor Artificial Intelligence, provides domain-optimized foundational capabilities for developing healthcare imaging training workflows in a native PyTorch paradigm. Useful for image pre-processing.

-
PyTorch: An open source machine learning framework that accelerates the path from research prototyping to production deployment. It takes care of the rest of the life hooks of the model.

3D Slicer: solve advanced image computing challenges with a focus on clinical and biomedical applications.
The software was extensively used during the data preparation hook. it is useful for converting NifTI files to DICOM and labeling (manually segmenting) the non-annotated data.
Depending on the size of the dataset, training could take up to days. Using GPUs helps reduce the training time, we could either use paid services like colab pro or make use of our own GPU thanks to cuda and cudnn.
pip install -r requirements.txt
Decathlon dataset, labeled
Liver Tumor Segmentation - Part 1, unlabeled
Liver Tumor Segmentation - Part 2, unlabeled