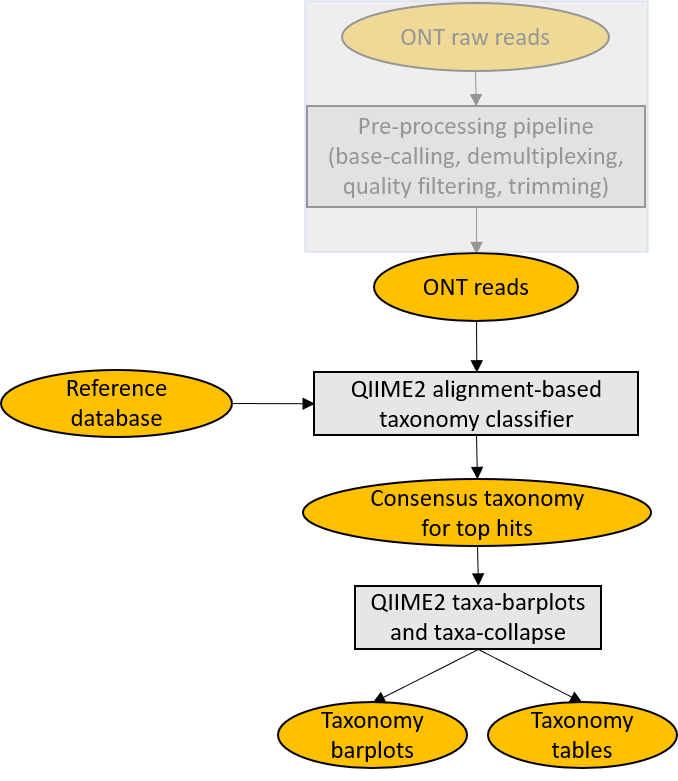
MetONTIIME is a Meta-barcoding pipeline for analysing ONT data in QIIME2 framework. The whole bioinformatic workflow consists of a preprocessing pipeline and a script emulating EPI2ME 16S workflow, aligning each read against a user-defined database, so to make the whole bioinformatic analysis from raw fast5 files to taxonomy assignments straightforward and simple. Tested with Ubuntu 20.04.1 LTS. For comparison of results obtained changing the reference database and PCR primers, have a look at Stephane Plaisance's interesting work.
Prerequisites
- Miniconda3.
Tested with conda 4.8.5.
which condashould return the path to the executable. If you don't have Miniconda3 installed, you could download and install it with:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
chmod 755 Miniconda3-latest-Linux-x86_64.sh
./Miniconda3-latest-Linux-x86_64.sh
Then, after completing MetONTIIME installation, set the MINICONDA_DIR variable in config_MinION_mobile_lab.R to the full path to miniconda3 directory.
- Guppy, the software for basecalling and demultiplexing provided by ONT. Tested with Guppy v6.0. If you don't have Guppy installed, choose an appropriate version and install it. For example, you could download and unpack the archive with:
wget /path/to/ont-guppy-cpu_version_of_interest.tar.gz
tar -xf ont-guppy-cpu_version_of_interest.tar.gz
A directory ont-guppy-cpu should have been created in your current directory. Then, after completing MetONTIIME installation, set the BASECALLER_DIR variable in config_MinION_mobile_lab.R to the full path to ont-guppy-cpu/bin directory.
- A fasta file downloaded from NCBI that you want to use as a reference database, or a preformatted marker gene reference database. For example, if you want to use the same database used by the EPI2ME 16S workflow for bacterial 16S gene, you can go to BioProject 33175, click send to, select Complete Record and File, set the Format to FASTA and then click Create File; the database can then be imported with Import_database.sh script after completing installation. In case you have downloaded a marker gene reference database instead, you already have sequence and taxonomy information in two separate text files. For example, if you want to download and import Silva_132_release database for 16S gene with sequences clustered at 99% identity, you can use the following instructions, after completing installation:
wget https://www.arb-silva.de/fileadmin/silva_databases/qiime/Silva_132_release.zip
unzip Silva_132_release.zip
source activate MetONTIIME_env
qiime tools import \
--type FeatureData[Sequence] \
--input-path SILVA_132_QIIME_release/rep_set/rep_set_16S_only/99/silva_132_99_16S.fna \
--output-path silva_132_99_16S_sequence.qza
qiime tools import \
--type FeatureData[Taxonomy] \
--input-path SILVA_132_QIIME_release/taxonomy/16S_only/99/taxonomy_7_levels.txt \
--input-format HeaderlessTSVTaxonomyFormat \
--output-path silva_132_99_16S_taxonomy.qza
Installation
git clone https://github.com/MaestSi/MetONTIIME.git
cd MetONTIIME
chmod 755 *
./install.sh
A conda environment named MetONTIIME_env is created, where seqtk, pycoQC, NanoFilt and qiime2-2021.8 are installed. Then, you can open the config_MinION_mobile_lab.R file with a text editor and set the variables PIPELINE_DIR and MINICONDA_DIR to the value suggested by the installation step.
The first time you run the MetONTIIME pipeline on a new database downloaded from NCBI, you can use the Import_database.sh script for importing a fasta file as a pair of QIIME2 artifacts. This script downloads some taxonomy files from NCBI (~9.4 GB) and uses entrez qiime and QIIME2 to generate a DNAFASTAFormat and a HeaderlessTSVTaxonomyFormat artifacts, containing sequences and corresponding taxonomy. Entrez_qiime is installed to a new conda environment named entrez_qiime_env. After this step, you can open the config_MinION_mobile_lab.R file with a text editor and set the variables DB and TAXONOMY to the newly generated QIIME2 artifacts. Both Blast and Vsearch aligners are supported and can be selected setting CLASSIFIER variable. After that, you can run the full MetONTIIME pipeline using the wrapper script Launch_MinION_mobile_lab.sh. The script Evaluate_diversity.sh can be used afterwards to generate a phylogenetic tree and compute some alpha and beta diversity metrics.
Import_database.sh
Usage: Import_database.sh <"sample_name".fasta>
Input:
- <"sample_name".fasta>: a fasta file downloaded from NCBI containing sequences that you want to use as a reference database
Outputs:
- <"sample_name"_sequence.qza>: QIIME2 artifact of type DNAFASTAFormat containing reference sequences
- <"sample_name"_taxonomy.qza>: QIIME2 artifact of type HeaderlessTSVTaxonomyFormat containing taxonomy of reference sequences
Launch_MinION_mobile_lab.sh
Usage: Launch_MinION_mobile_lab.sh <fast5_dir>
Input:
- <fast5_dir>: directory containing raw fast5 files
Outputs (saved in <fast5_dir>_analysis/analysis):
- feature-table_absfreq.tsv, feature-table_absfreq_level$lev.tsv: files containing the number of reads assigned to each taxa for each sample, collapsed at different taxonomic levels
- feature-table_relfreq.tsv, feature-table_relfreq_level$lev.tsv: files containing the proportion of reads assigned to each taxa for each sample, collapsed at different taxonomic levels
- taxa-bar-plots.qzv: QIIME2 visualization artifact of barplots with taxonomy abundances
- taxa-bar-plots-no-Unassigned.qzv: QIIME2 visualization artifact of barplots with taxonomy abundances excluding Unassigned reads
- demux_summary.qzv: QIIME2 visualization artifact with summary of sequences assigned to each sample after demultiplexing
- logfile.txt, manifest.txt, sequences.qza, table.qz*, rep-seqs.qz*, taxonomy.qz*, table_collapsed.qza, feature-table_absfreq.biom, table_collapsed_relfreq.qz*, feature-table_relfreq.biom: temporary files useful for debugging or for further analyses
Outputs (saved in <fast5_dir>_analysis/qc):
- Read length distributions and pycoQC report
Outputs (saved in <fast5_dir>_analysis/basecalling):
- Temporary files for basecalling
Outputs (saved in <fast5_dir>_analysis/preprocessing):
- Temporary files for demultiplexing, filtering based on read length and adapters trimming
Evaluate_diversity_non_phylogenetic.sh
Usage: Evaluate_diversity_non_phylogenetic.sh -f <feature_table> -m <sample_metadata> -d <sampling_depth>
Note: can be run in background with nohup; the script reads a feature table collapsed at a desired taxonomic level (e.g. genus), subsamples the same number of reads for each sample and computes some alpha and beta non-pyhlogenetic diversity metrics.
Inputs:
- <feature_table>: feature table collapsed at a desired taxonomic level (e.g. $WORKING_DIRECTORY/collapsed_feature_tables/table_collapsed_absfreq_level5.qza)
- <sample_metadata>: file containing meta-data for samples, generated by MetONTIIME.sh if not provided by the user
- <sampling_depth>: number of reads subsampled for each sample for normalizing the collapsed feature table; this value can be chosen looking at demu_summary.qzv or at logfile.txt
Outputs:
- core-metrics-results_<feature_table>_<sampling_depth>_subsampled_non_phylogenetic: folder containing some alpha and beta diversity metrics
- alpha-rarefaction_<feature_table>_<sampling_depth>_subsampled_non_phylogenetic.qzv: visualization artifact describing alpha diversity as a function of sampling depth
Evaluate_diversity.sh
Usage: Evaluate_diversity.sh -w <working_directory> -m <sample_metadata> -d <sampling_depth> -t <threads> -c <clustering_threshold>
Note: can be run in background with nohup; this script is experimental, suggestions for improving the logic behind it are welcome; the script subsamples the same number of reads for each sample, performs clustering at <clustering_threshold> threshold and considers these to be the representative sequences. It then uses representative sequences for building a phylogenetic tree and for computing some alpha and beta diversity metrics.
Inputs:
- <working_directory>: directory containing rep-seqs.qza and table.qza artifacts generated by MetONTIIME.sh and fastq.gz files
- <sample_metadata>: file containing meta-data for samples, generated by MetONTIIME.sh if not provided by the user
- <sampling_depth>: number of reads subsampled for each sample before clustering; this value can be chosen looking at demu_summary.qzv or at logfile.txt
- <threads>: number of threads used for generating the phylogenetic tree
- <clustering_threshold>: clustering similarity threshold in (0, 1] used for picking representative sequences
Outputs:
- core-metrics-results_<sampling_depth>_subsampled: folder containing some alpha and beta diversity metrics
- alpha-rarefaction_<sampling_depth>_subsampled.qzv: visualization artifact describing alpha diversity as a function of sampling depth
- <"sample_name">_<sampling_depth>_subsampled.fastq.gz, manifest_<sampling_depth>_subsampled.txt, aligned-repseqs_<sampling_depth>_subsampled.qza, masked-aligned-rep-seqs_<sampling_depth>_subsampled.qza, rooted-tree_<sampling_depth>_subsampled.qza, unrooted-tree_<sampling_depth>_subsampled.qza: temporary files generated for calculating diversity metrics
In case you have already performed basecalling, demultiplexing, quality filtering, adapters and PCR primers trimming, and already have BC<num>.fastq.gz files, you could run the pipeline using the following instruction. All parameters are required with no default values.
source activate MetONTIIME_env
nohup ./MetONTIIME.sh [-w working_dir] [-f metadata_file] [-s sequences_artifact] [-t taxonomy_artifact] [-n num_threads] [-c taxonomic_classifier] [-m max_accepts] [-q min_query_coverage] [-i min_id_thr] &
where:
<working_dir>: full path to directory containing fastq.gz files
<metadata_file>: full path to metadata file; if the file doesn't exist yet, it is created by the pipeline
<sequences_artifact>: full path to <file name>_sequence.qza QIIME2 artifact, may be created by Import_database.sh script
<taxonomy_artifact>: full path to <file name>_taxonomy.qza QIIME2 artifact, may be created by Import_database.sh script
<num_threads>: maximum number of threads used
<taxonomic_classifier>: either Blast or Vsearch
<max_accepts>: maximum number of hits; if a value > 1 is used, a consensus taxonomy for the top hits is retrieved
<min_query_coverage>: minimum portion of a query sequence that should be aligned to a sequence in the database [0-1]
<min_id_thr>: minimum alignment identity threshold [0-1]
In the following, auxiliary scripts run by Launch_MinION_mobile_lab.sh are listed. These scripts should not be called directly.
MinION_mobile_lab.R
Note: script run by Launch_MinION_mobile_lab.sh.
config_MinION_mobile_lab.R
Note: configuration script, must be modified before running Launch_MinION_mobile_lab.sh.
MetONTIIME.sh
Note: script run by MinION_mobile_lab.R for performing taxonomic classification in QIIME2 framework.
subsample_fast5.sh
Note: script run by MinION_mobile_lab.R if do_subsampling_flag variable is set to 1 in config_MinION_mobile_lab.R.
All .qzv and .qza artifacts can be visualized either importing them to QIIME2 View or with command:
source activate MetONTIIME_env
qiime tools view <file.qz*>
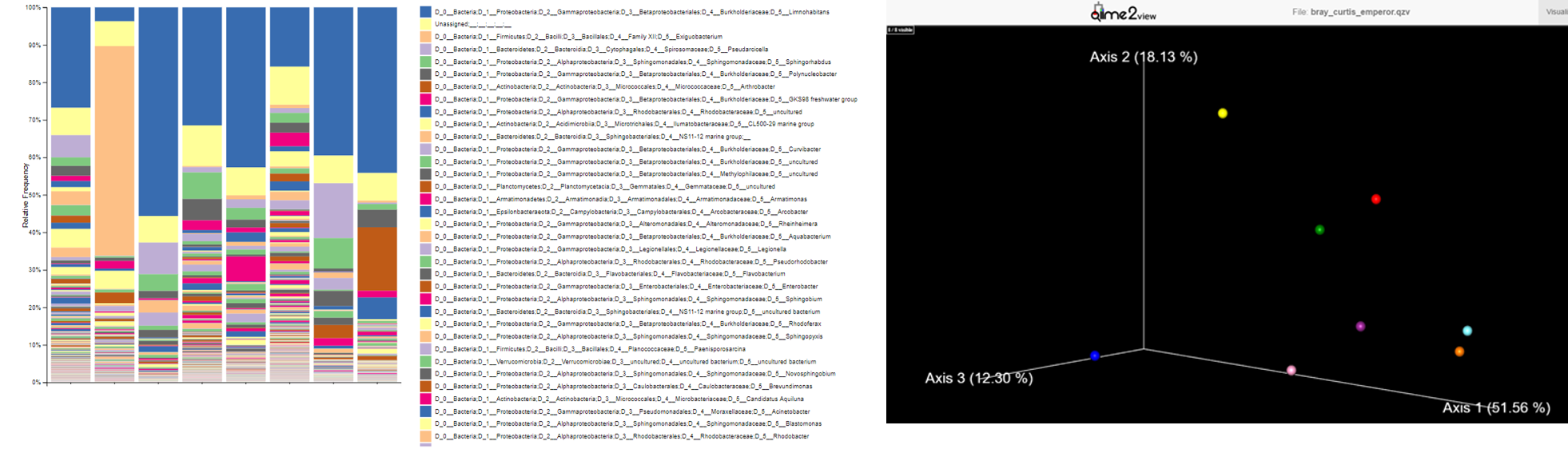
In particular, you could visualize an interactive multi-sample taxonomy barplot, describing the composition of each sample at the desired taxonomic level, and a PCA plot of Beta-diversity among samples.
The MetONTIIME pipeline is composed of a preprocessing pipeline inherited from ONTrack and of some wrapper scripts for QIIME2 and entrez qiime.
For further information and insights into pipeline development, please have a look at my doctoral thesis.
Maestri, S (2021). Development of novel bioinformatic pipelines for MinION-based DNA barcoding (Doctoral thesis, Università degli Studi di Verona, Verona, Italy). Retrieved from https://iris.univr.it/retrieve/handle/11562/1042782/205364/.
Please, refer to the following manuscripts for further information.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, and Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37: 852–857. https://doi.org/10.1038/s41587-019-0209-9
Baker CCM (2016). entrez qiime: a utility for generating QIIME input files from the NCBI databases. github.com/bakerccm/entrez qiime, release v2.0, 7 October 2016. doi:10.5281/zenodo.159607
Maestri S, Cosentino E, Paterno M, Freitag H, Garces JM, Marcolungo L, Alfano M, Njunjić I, Schilthuizen M, Slik F, Menegon M, Rossato M, Delledonne M. A Rapid and Accurate MinION-Based Workflow for Tracking Species Biodiversity in the Field. Genes. 2019; 10(6):468. https://doi.org/10.3390/genes10060468