current version 0.1.1 Maintained branch: hpc_version
Our lab uses this program for cell cycle analysis in high content screening format using data on our omero server. Additional bug fixes and testing necessary! Requires a connection to an active Omero server
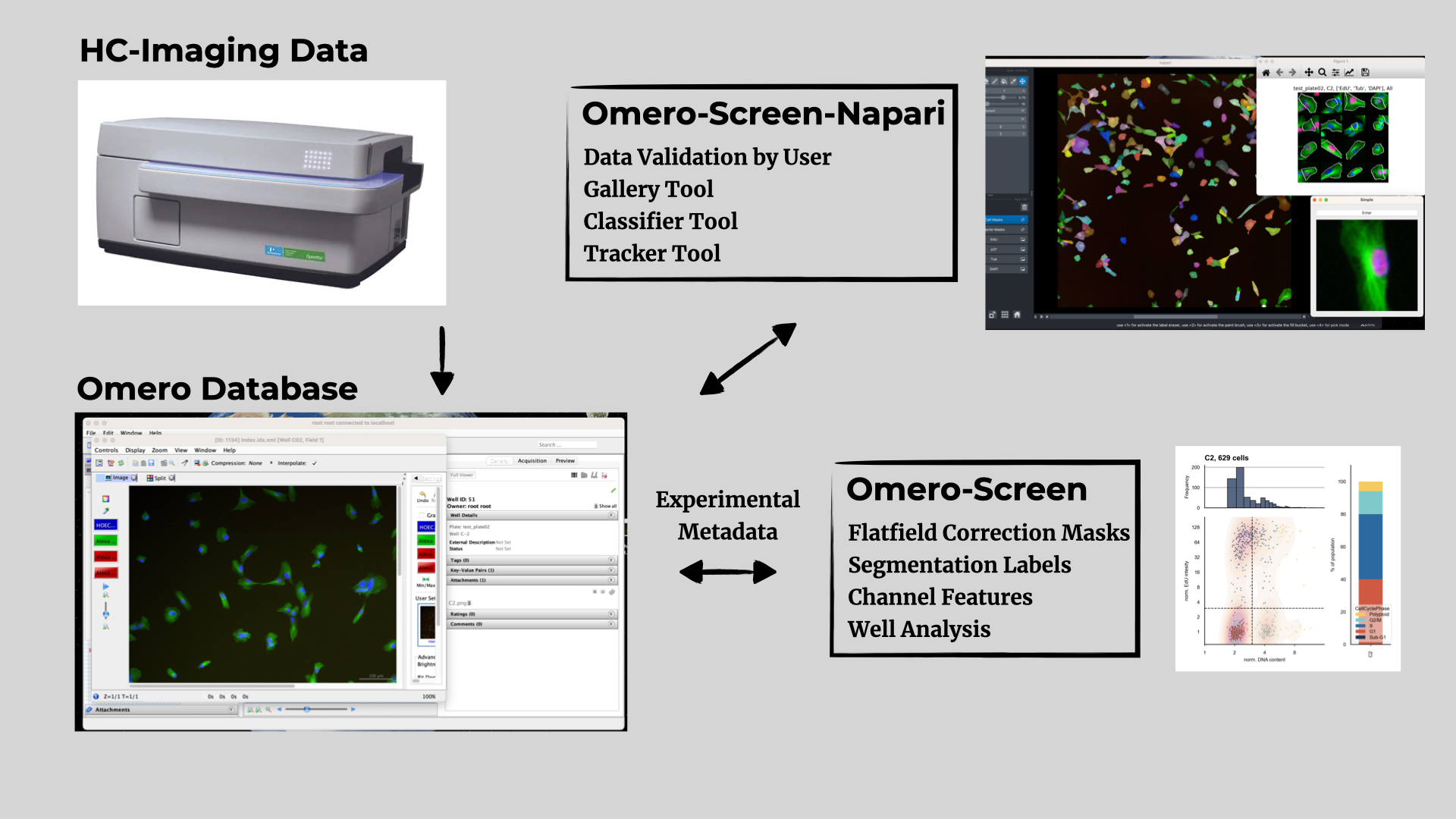
An outline of the current workflow is shown below:
Robert Zach, Haoran Yue, Alex Herbert and Helfrid Hochegger
An end to end pipeline to analyse high-content immuno-fluorescence data. the software is designed to work with data stored on an Omero server and handles experimental metadata, flatfield correction, image segmentation and cell cycle analysis, if EdU labelling was performed. A separate Napari plugin Omero-Screen-Napari is available to display the data in Napari and provides further functionality to interact with the data.
We have not yet packaged the project for pip installation. To install the package, clone the repository and install the requirements. The program can be run via bin/omero_screen_run_term via the terminal or via the main.py script in the root directory.