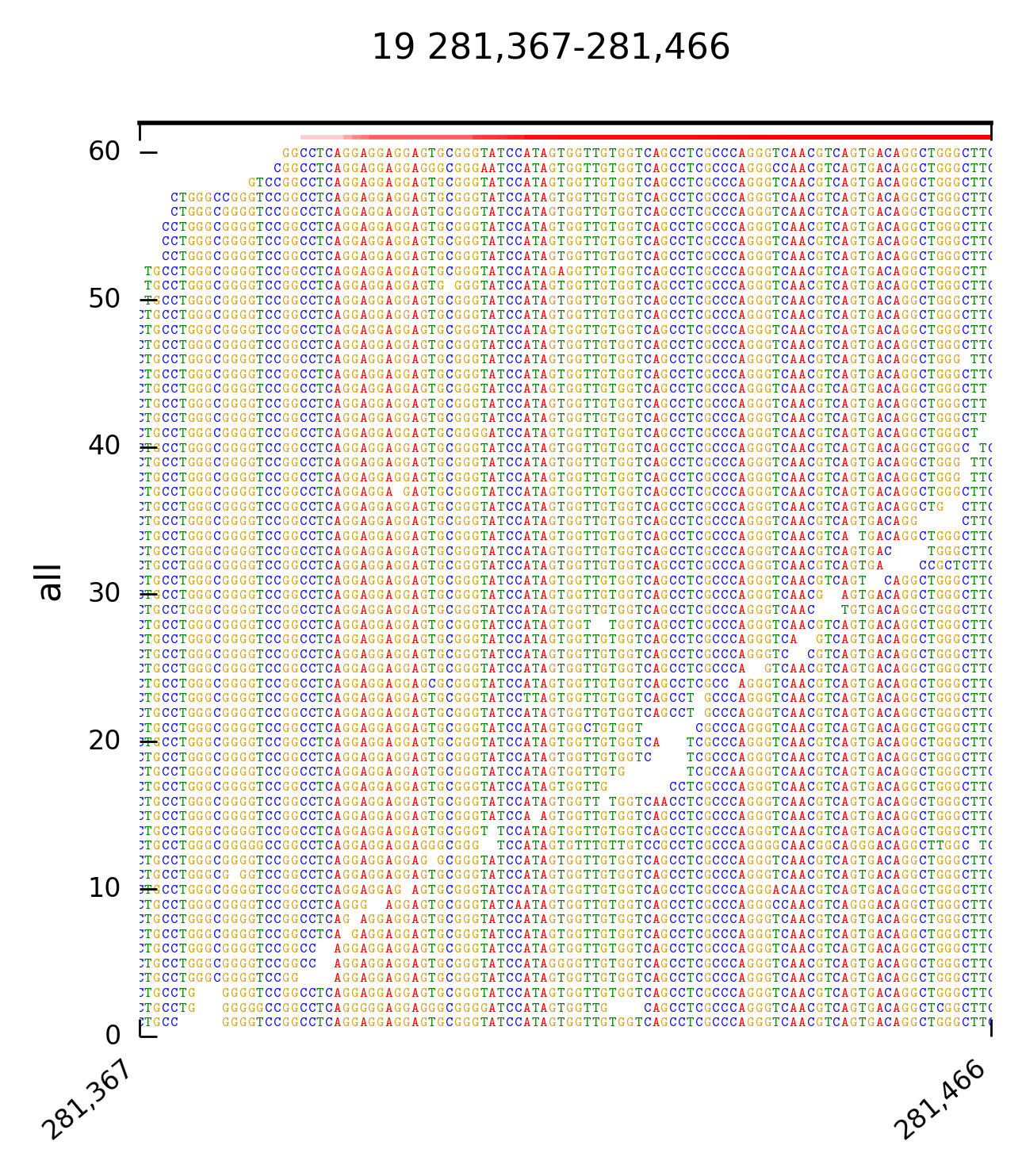
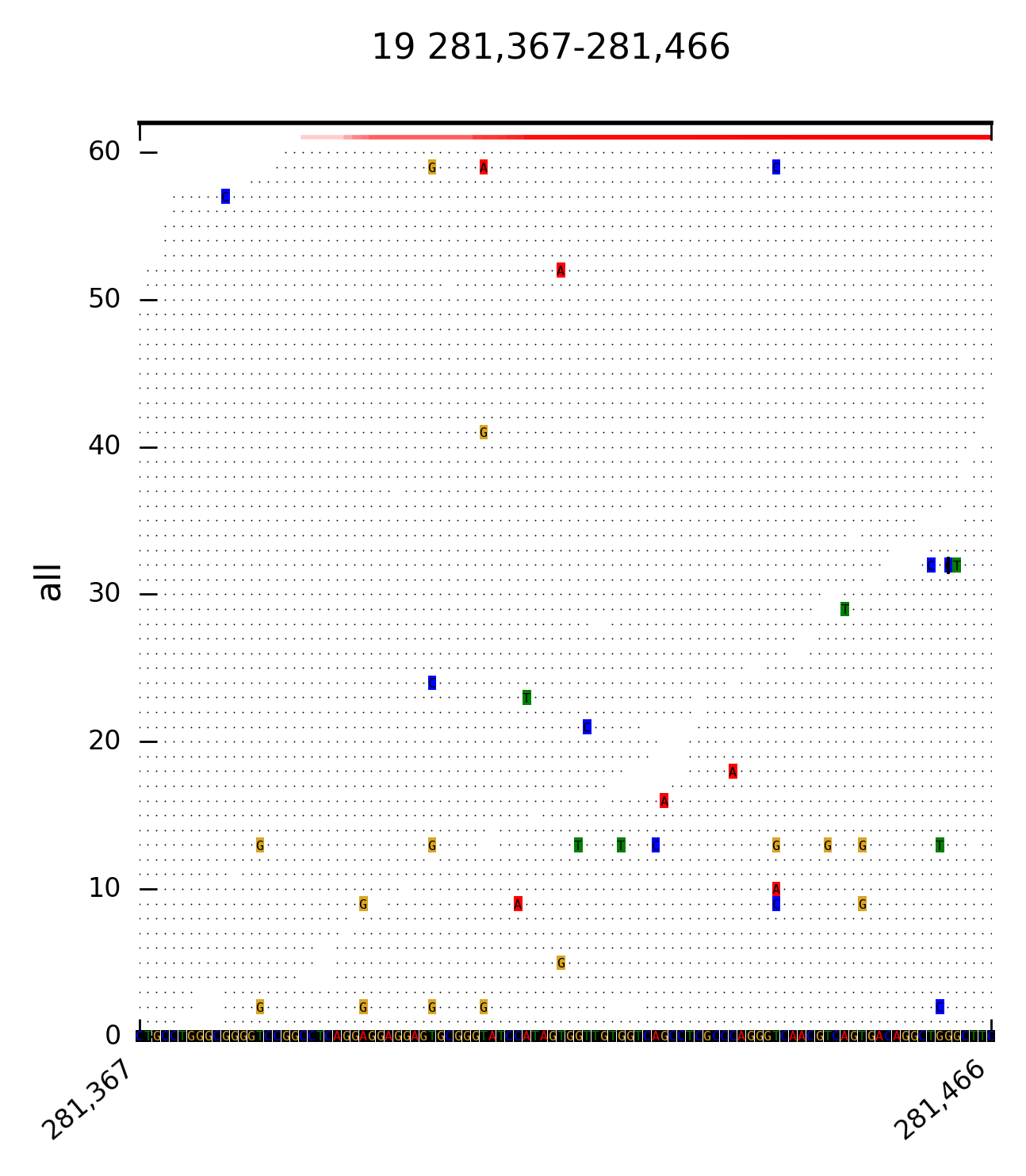
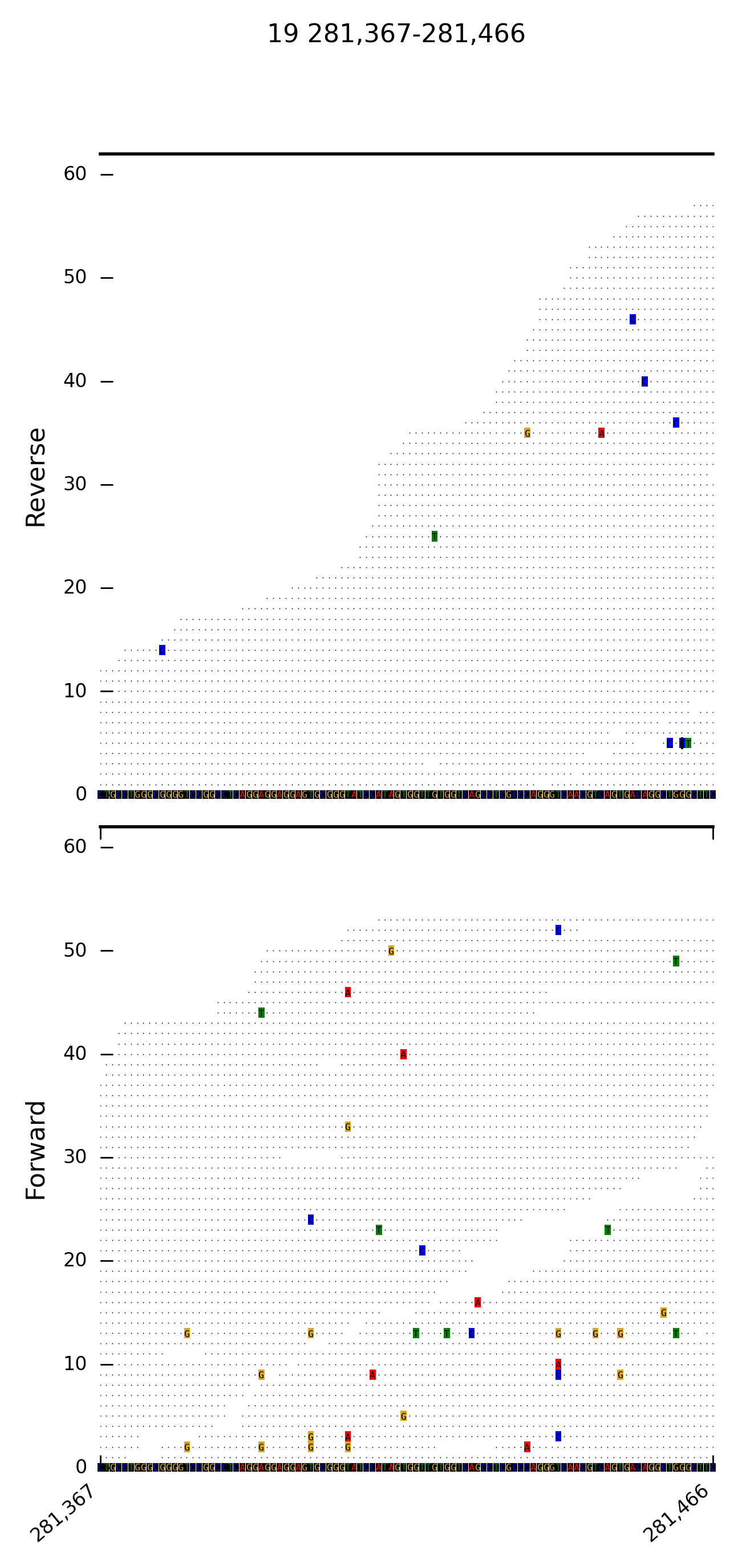
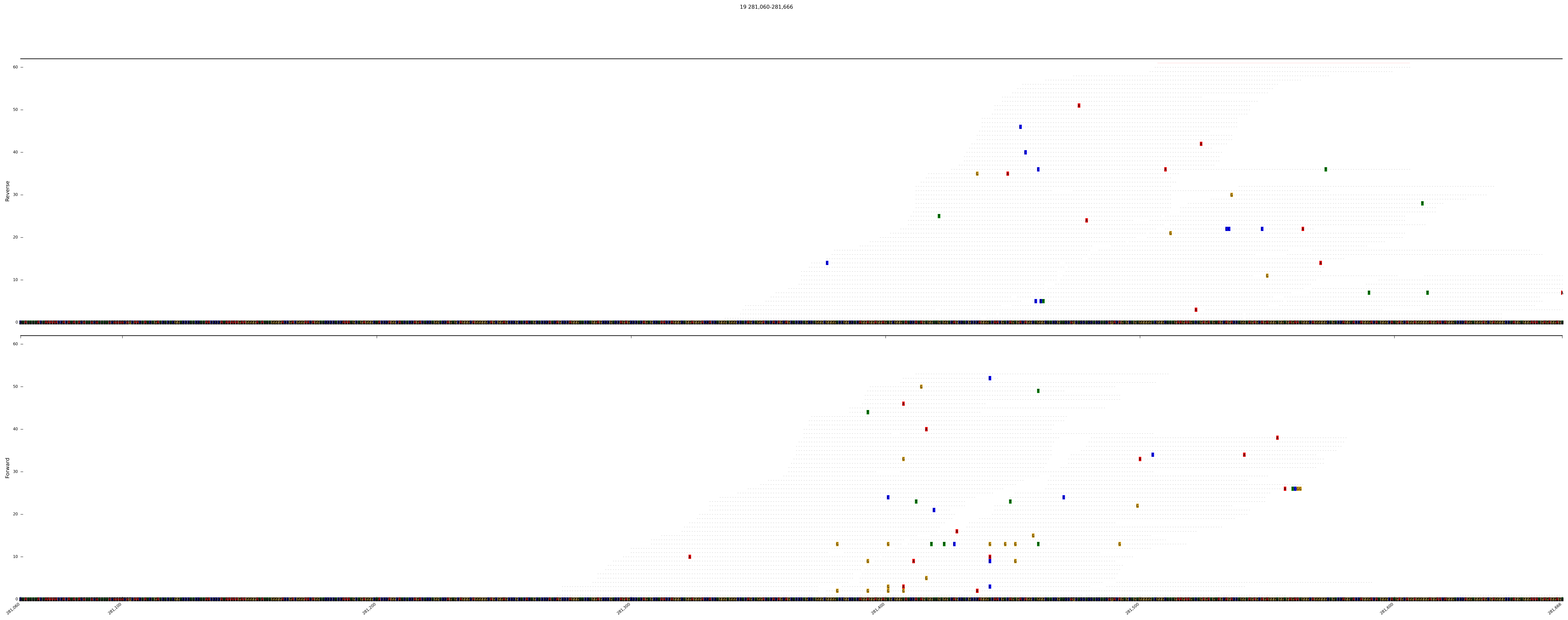
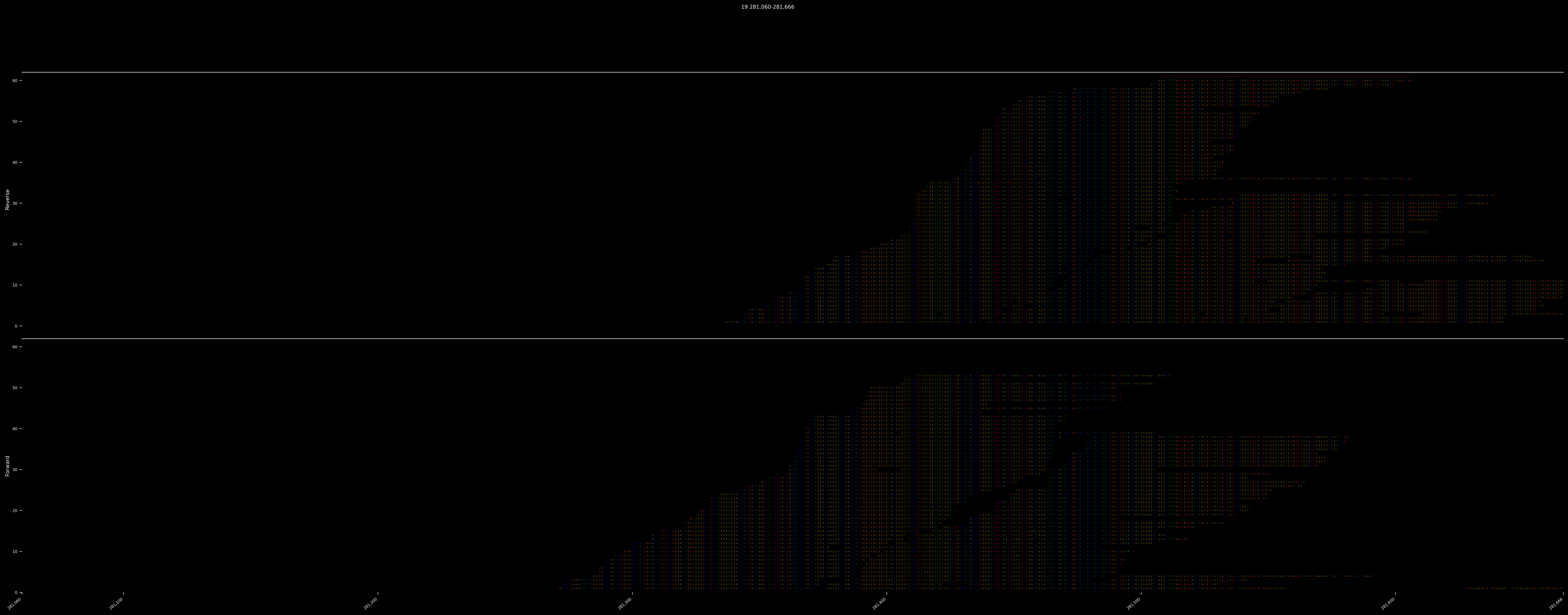
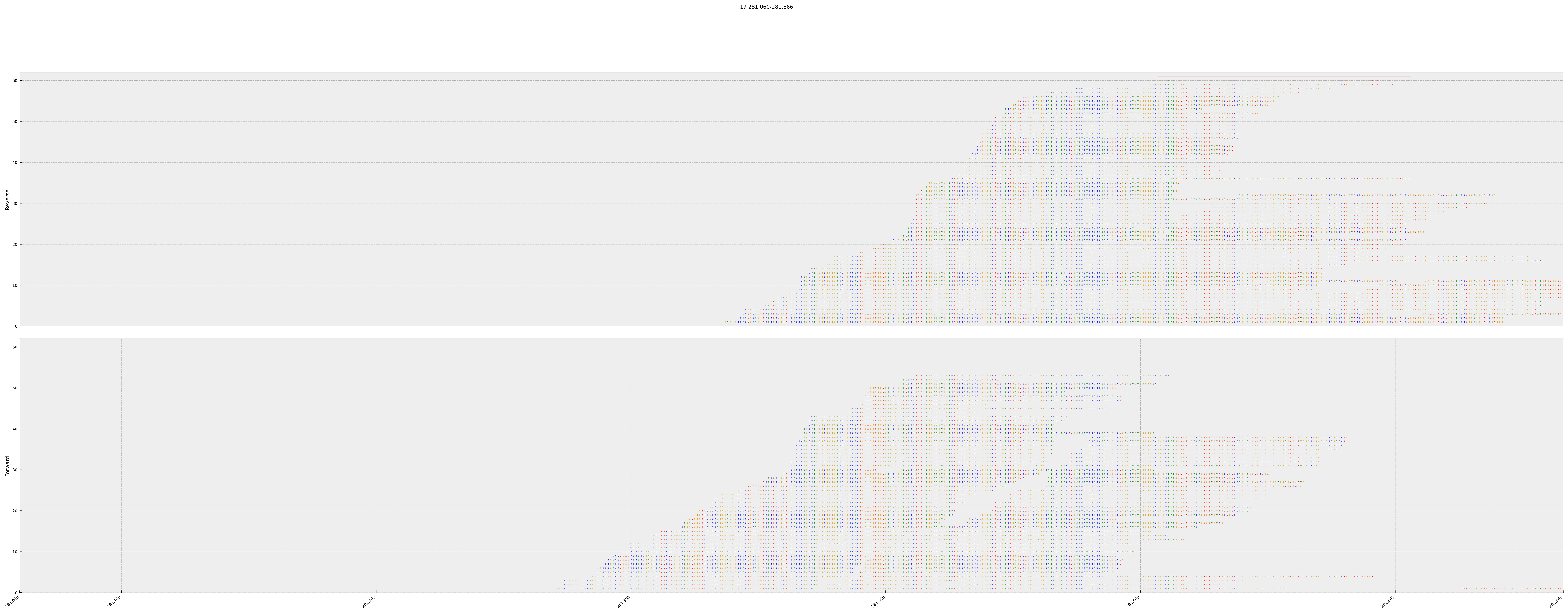
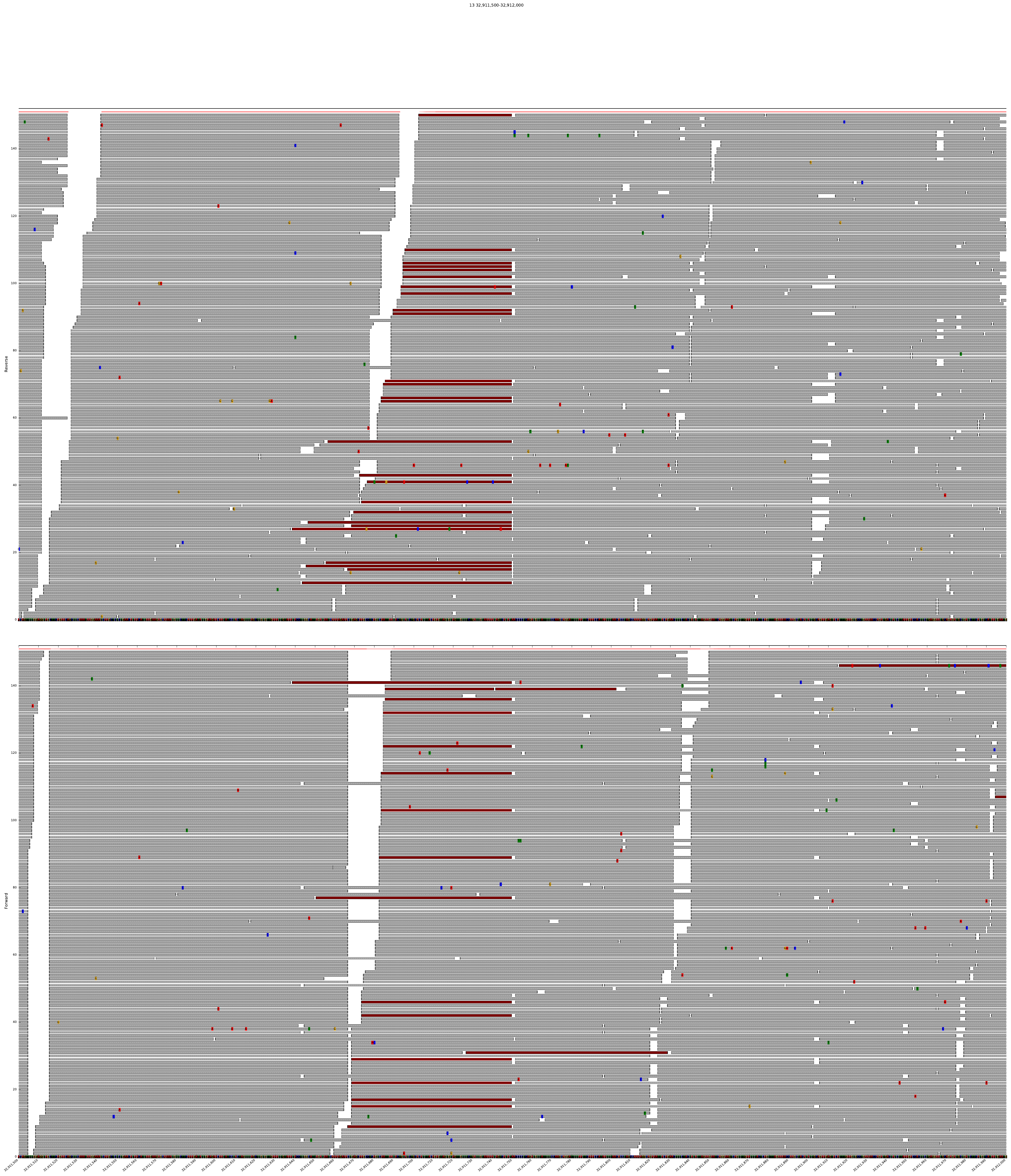
DrukBam can be used with or without a reference fasta file and allows fast plotting multiple variants or regions of interest. Please provide feedback like bugs or options you might miss, I wrote this programm because I did not found a convicning tool to provide fast plotting of alignemnts withput using a GUI like in IGV or Tablet.
- pysam
- pandas
- matplotlib
- tqdm
DrukBam is available via pypi:
pip install drukbam==1.1.4
docker image
docker pull stephanholgerdrukewitz/drukbam:1.1.4
docker usage
docker run -it --rm -v $PWD:/data drukbam:1.1.4 DrukBam region -s 281367 -e 281468 -c 19 -b /data/test_data/test_small.bam --outfmt png -i example_out_small --maxcoverage 60 --outlineoff
❗ ❗ versions <1.1.4 are deprecated and should not be used anymore ❗ ❗
DrukBam vcf
usage: DrukBam vcf [-h] -b BAM -v VCF [-p PADDING] [--highlight]
[--threads THREADS] [--maxcoverage MAXCOVERAGE]
[--direction] [--schematic] [--style STYLE] [--fasta FASTA]
[--outputdir OUTPUTDIR] [-i ID] [--chunksize CHUNKSIZE]
[--outfmt OUTFMT] [--outlineoff]
optional arguments:
-h, --help show this help message and exit
required arguments:
-b BAM, --bam BAM Pos. sorted and indexed bam file
-v VCF, --vcf VCF vcf file with variants of interest
-p PADDING, --padding PADDING
number of nt around the variant
--highlight highlight the position of interest
optional arguments:
--threads THREADS number of cpu's to run in paralell, ROI <1000 will
always use 1 core
--maxcoverage MAXCOVERAGE
max cov to plot
--direction split reads by forward and reverse
--schematic plot no nucleotide, recommended for ROI>1000
--style STYLE different style options for the plot, provide .ini
file
--fasta FASTA fasta file for reference related plotting
--outputdir OUTPUTDIR
directory for output
-i ID, --id ID output filename
--chunksize CHUNKSIZE
max size of visualized area, can be increases but will
sow down calculation
--outfmt OUTFMT format of plot, choose between pdf,svg,png
--outlineoff plotting of read outline
DrukBam region
usage: DrukBam region [-h] -b BAM -c CHROMOSOME -s START -e END
[--threads THREADS] [--maxcoverage MAXCOVERAGE]
[--direction] [--schematic] [--style STYLE]
[--fasta FASTA] [--outputdir OUTPUTDIR] [-i ID]
[--chunksize CHUNKSIZE] [--outfmt OUTFMT] [--outlineoff]
optional arguments:
-h, --help show this help message and exit
required arguments:
-b BAM, --bam BAM Pos. sorted and indexed bam file
-c CHROMOSOME, --chromosome CHROMOSOME
name of chromosome/contig
-s START, --start START
start of region of interest
-e END, --end END end of the region of interest
optional arguments:
--threads THREADS number of cpu's to run in paralell, ROI <1000 will
always use 1 core
--maxcoverage MAXCOVERAGE
max cov to plot
--direction split reads by forward and reverse
--schematic plot no nucleotide, recommended for ROI>1000
--style STYLE different style options for the plot, provide .ini
file
--fasta FASTA fasta file for reference related plotting
--outputdir OUTPUTDIR
directory for output
-i ID, --id ID output filename
--chunksize CHUNKSIZE
max size of visualized area, can be increases but will
sow down calculation
--outfmt OUTFMT format of plot, choose between pdf,svg,png
--outlineoff plotting of read outline
The following command will create an image of that region:
DrukBam region -s 281367 -e 281468 -c 19 -b test_data/test_small.bam --outfmt png -i example_out --maxcoverage 60 --outlineoff --fasta test_data/chr19_first500k.fasta
The arguments used above are:
-s start of ROI
-e end of ROI
-c chromosome of ROI
-b alignment file, sorted and indexed
--outfmt format of plot
-i ID which is used for naming the plot
--maxcoverage yaxis max of plot
--outlineoff dont draw outlines around every read
--fasta location of ref. fasta
The following command will plot all positions in a vcf file:
DrukBam vcf -b test_data/test_small.bam -v example.vcf --padding 100 -i example_vcf --maxcoverage 60 --fasta test_data/chr19_first500k.fasta --threads 12
The arguments used above are:
-b alignment file, sorted and indexed
-v vcf file
-i ID which is used for naming the plot
--maxcoverage yaxis max of plot
--fasta location of ref. fasta
--threads number of cpu's to use,
- color and style can be changed using the --style option and providing a style.ini file
- official matplotlib colors are allowed color list
- pltstyle can be changed style list