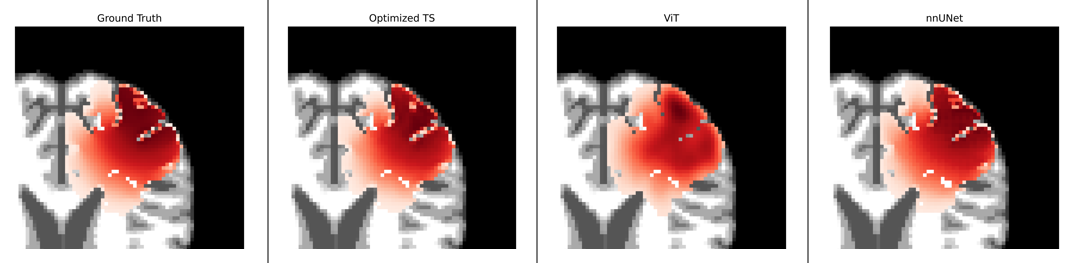
TumorNetSolvers is an adaptive deep learning framework designed for efficient personalization of PDE-based tumor growth models, focused on simulating and predicting tumor evolution. Built on the powerful nnU-Net framework, it leverages state-of-the-art neural architectures to address both forward and inverse tumor modeling problems.
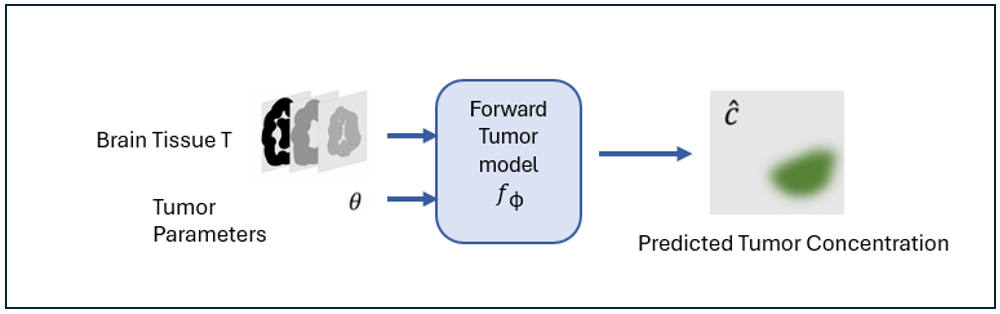
TumorNetSolvers provides a flexible and efficient solution for simulating tumor growth in 3D using Partial Differential Equations (PDEs), specifically the Fisher-Kolmogorov equation. It personalizes the model for individual patients by inferring tumor growth parameters (diffusion coefficient, growth rate, initial location) from MRI scans or similar medical images.
- Input: A 3D medical image (e.g., brain tissue segmentations) and tumor parameters (growth rate, diffusion coefficient, tumor location).
- Output: A 3D simulation of tumor corresponding to the given anatomy and tumor parameters.
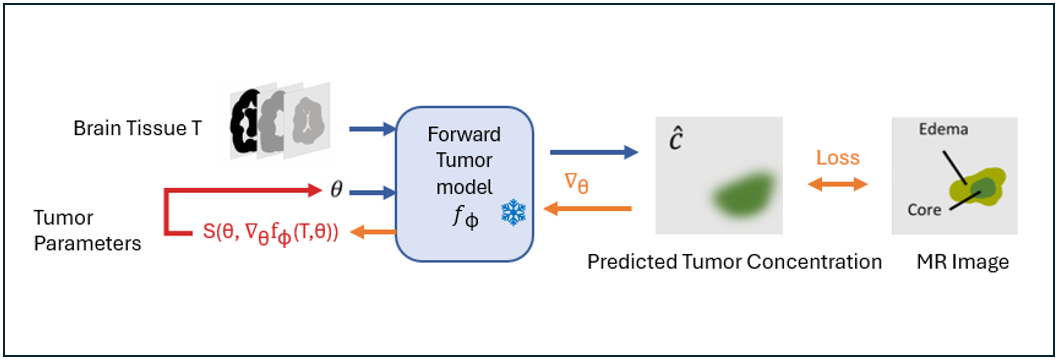
Given a high-performing differentiable forward model, we "freeze" the model and optimize its parameters based on a target tumor simulation.
- Input: A target tumor simulation (e.g., MRI-based 3D tumor image).
- Goal: Infer the underlying tumor parameters (e.g., growth rate, diffusion coefficient, tumor location) by adjusting these input parameters to match the target simulation.
TumorNetSolvers is trained on the Fisher-Kolmogorov equation to model brain tumor cell dynamics over time and space:
Where:
- ( c(x, t) ) is the tumor cell concentration,
- ( D ) is the diffusion coefficient,
- ( \rho ) is the growth rate,
- ( \nabla \cdot (D \nabla c) ) is the diffusion term,
- ( \rho c(1 - c) ) models logistic growth, limiting ( c ) as it approaches 1.
TumorNetSolvers uses deep learning to approximate this PDE's solutions but can, in theory, adapt to different PDE-based models.
Neural networks approximate the PDE solution, mapping inputs (image + parameters) to outputs (simulation).
The framework builds upon nnU-Net's self-configuring capabilities, adapting it for conditioned image-to-image regression tasks.
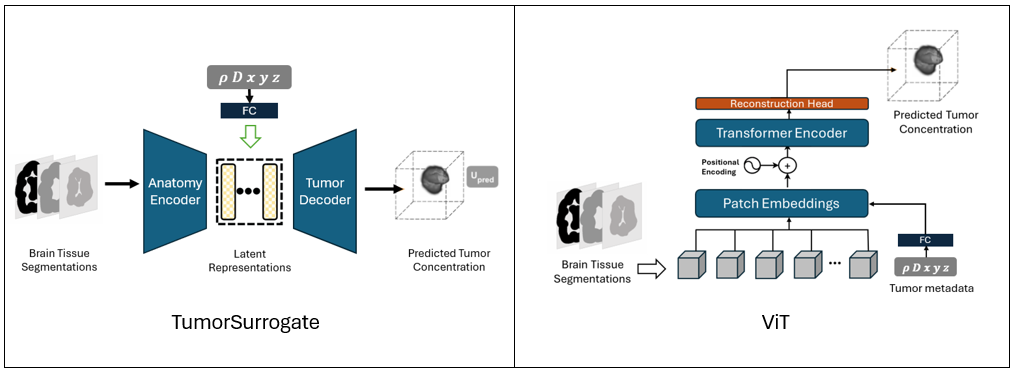
TumorNetSolvers includes three key models:
-
TumorSurrogate (Baseline)
- A 3D CNN that integrates anatomy info and tumor metadata for conditioned tumor simulation.
-
Modified nnU-Net
- An adapted version of the powerful self-configuring nnU-Net framework for tumor growth simulation using conditioned inputs.
-
3D Vision Transformer (ViT)
- A transformer-based model incorporating tumor metadata as additional input tokens in the embedding space.
For detailed usage, refer to the scripts in the scripts directory or follow the instructions below:
-
Forward Problem
Run the scriptrunning_inference.pywith the necessary inputs (3D medical image, tumor parameters) to generate tumor simulations. -
Inverse Problem
Run the scriptpredict_tumor_params.pyto infer tumor parameters by optimizing against a target tumor simulation.
To get started with TumorNetSolvers, install it directly from GitHub by following these steps:
-
Clone the repository:
git clone https://github.com/ZeinebZH/tumornetsolvers.git cd tumornetsolvers -
Install the required dependencies and the package:
pip install -r requirements.txt pip install .
We welcome contributions to TumorNetSolvers!
This project is licensed under the MIT License and incorporates code from nnU-Net, licensed under Apache 2.0. Both licenses apply and must be respected. See the LICENSE file for details.
- Weidner J, et al. Rapid Personalization of PDE-Based Tumor Growth Using a Differentiable Forward Model. Medical Imaging with Deep Learning, 2024. DOI
- Ezhov I, et al. Geometry-aware Neural Solver for Fast Bayesian Calibration of Brain Tumor Models. IEEE Transactions on Medical Imaging, 2021;41(5):1269–78. DOI
- Isensee F, et al. nnU-Net: A Self-configuring Method for Deep Learning-based Biomedical Image Segmentation. Nature Methods, 2021;18(2):203–11. DOI
- Lin K, Heckel R. Vision Transformers Enable Fast and Robust Accelerated MRI. International Conference on Medical Imaging with Deep Learning, 2022. DOI