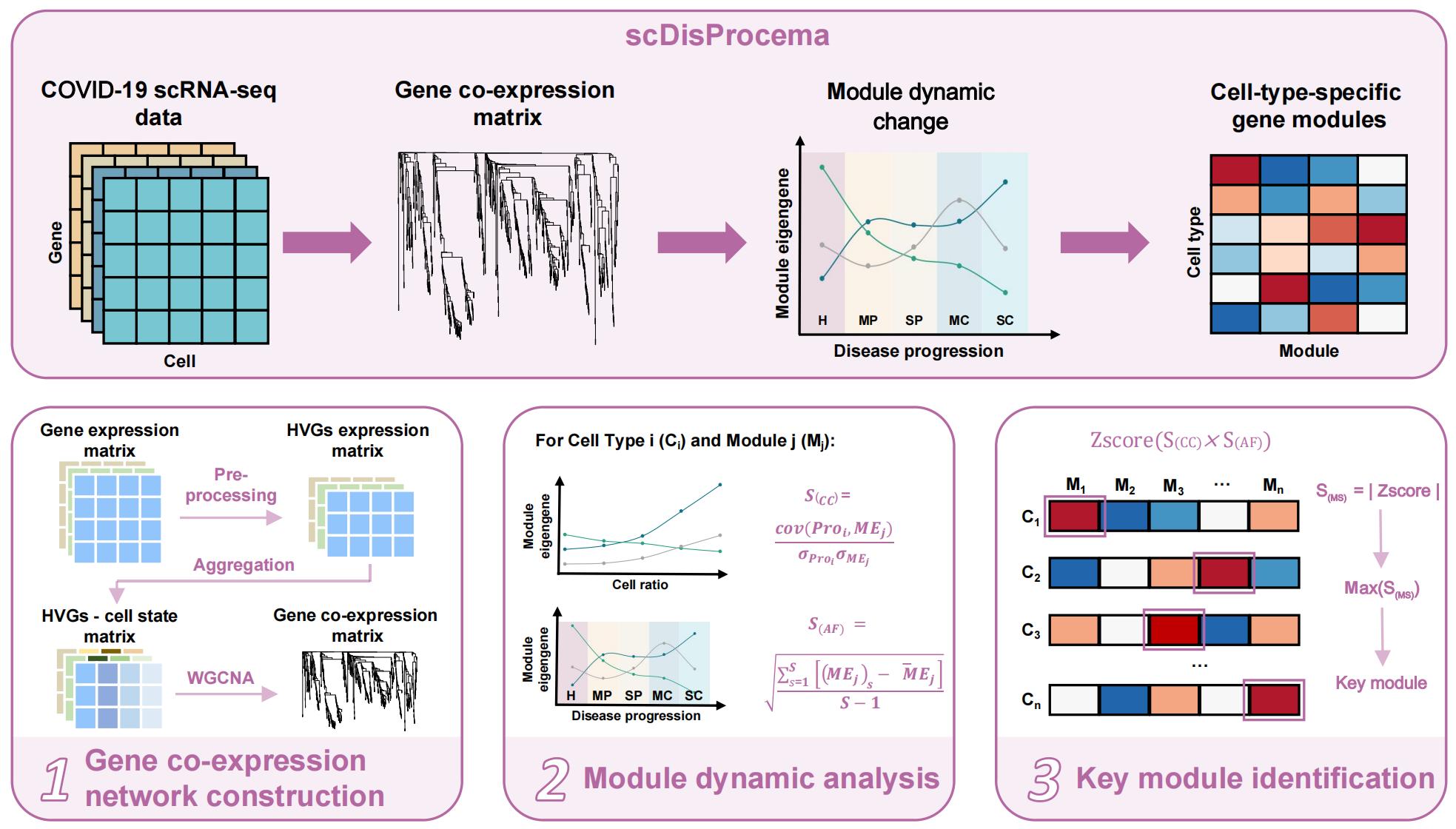
A GCN construction pipeline to trace dynamic changes of immune system response during disease progression using single-cell data
Varying degrees of dynamic changes could occur in many aspects over disease progression and recovery process , such as cell proportion, clinical traits, or gene expression pattern. And there may be significant correlation between changes in cell states and gene modules. We proposed the scDisProcema, namely single-cell Disease Progression cellular module analysis, to capture this dynamic change correlation. First we inferred gene co-expression network modules during disease progression. Then we identified key modules underlying the dynamic changes of regulation in disease processes by correlation module expression patterns with changes of the traits or proportion of cell types. The underlying biological processes can be elucidated by further researches of the hub genes or functions of the key modules. The above steps traced the dynamic changes during the disease development and revealed the gene expression patterns corresponding to these cellular changes.
please download scDisProcema_0.1.0.tar.gz from the release page and execute the following command
# download the source package of scDisProcema-1.0.tar.gz and install it
# ensure the right directory for scDisProcema-1.0.tar.gz
# ensure the dependency packages have been installed.('WGCNA', 'Seurat', 'stringr', 'stats', 'magrittr', 'dplyr', 'Hmisc','dynamicTreeCut','utils')
install.packages(pkgs = 'scDisProcema-1.0.tar.gz',repos = NULL, type = "source")
Please refer to the tutorial vignette.
scDisProcema repository was developed by Anyao Li and Jihong Yang. Should you have any questions, please contact us at lianyao625@163.com.