The IGV-Web app is a pure-client "genome browser" application based on igv.js. It is developed by the Integrative Genomics Viewer (IGV) team. You can use our hosted app at https://igv.org/app, or follow the directions below to install your own.
Note: the instructions below are for developers or others wishing to install a copy of the IGV-Web app. For user documentation see https://igvteam.github.io/igv-webapp/. A link to the user documentation is also provided in the app's Help menu.
The IGV-Web app and igv.js require a modern web browser with support for JavaScript ECMAScript 2015.
IGV-Web is a pure client web application consisting entirely of static files and can be served from virtually any web server, e.g. Apache, Flask, Tomcat, nginx, or Amazon S3. You can install it from a pre-built distribution package or by building it from source code. To run the app, refer to your web server documentation for information on serving web content. Some examples are provided in the section on Running the app below.
A zip file containing the pre-built app can be downloaded from https://igv.org/app-archive.
-
Requirements
- Node >= v8.11.4
- NPM >= v5.6.0
-
Get the source code either (a) by downloading a zip file via the green
Codebutton at https://github.com/igvteam/igv-webapp, or (b) by cloning the repository:
git clone https://github.com/igvteam/igv-webapp.git
- Change directories to project
cd ./igv-webapp
- Install
npm install
- Build
npm run build
As noted above, refer to your web server documentation for instructions on serving the web content. As examples, instructions for use with the NPM package http-server and Amazon S3 follow.
For more information about http-server, including installing it locally, see the http-server documentation.
- Start http-server on the web content directory (i.e., the downloaded pre-packaged distribution described above, or the
distdirectory if building from source )
npx http-server -a localhost <web content directory>
- Open a browser and enter the following
localhost:8080
or
localhost:8080/index.html
To serve the app from Amazon S3, simply upload the contents from the IGV-Web content directory (i.e., the pre-packaged distribution described above, or the dist directory if building from source ) to an Amazon S3 bucket. Keep the
directory structure intact, and make all files public. Example of an S3 hosted app is at
https://s3.amazonaws.com/igv.org.app/app/index.html. Note this is an example and is not kept up-to-date.
NOTE:
- If you installed a pre-built distribution package, the configuration files referred to below can be found in the
resourcesdirectory. - In the the source code, you will find the configuration files in the
resourcesdirectory, but to deploy any changes you make there, you must rebuild thedistdirectory withnpm run build. Alternatively, you can just directly update files indist/resources. Note however, any changes to files indistwill be overwritten the next time you do a build.
The IGV-Web app is configured with the global igvwebConfig defined in igvwebConfig.js. The following properties are customizable.
-
genomes- URL to a JSON file containing a list of genome configuration objects. This list populates the Genomes pulldown menu. For a description of the genome configuration object, see the igv.js wiki . -
trackRegistryFile- URL to a configuration file for the Tracks pulldown menu. Use this to define custom track menus. The file contains a JSON object with genomeIDs as keys and an array of URLs to JSON files defining menu entries in the Tracks pulldown menu. The default track registry file isresources/tracks/trackRegistry.json. Further details on configuring the tracks menu are available below. -
igvConfig- An igv.js configuration object. See the igv.js wiki for details. -
clientId- (optional) A Google clientId, used to enable OAuth for the Google picker and access to protected Google resources. See Google Support for instructions on obtaining a clienId. OAuth requests from igv.js will include the following scopes.- https://www.googleapis.com/auth/userinfo.profile - always
- https://www.googleapis.com/auth/devstorage.read_only - if accessing protected Google Cloud Storage files
- https://www.googleapis.com/auth/drive.readonly' - if accessing files stored on Google Drive
-
urlShortener- An object or function defining a URL shortener to shorten links created by the Share button. The value of this property can be replaced with a function, taking a single argument (the long URL) and returning the shortened URL, or an Object.
var igvwebConfig = {
genomes: "https://s3.amazonaws.com/igv.org.genomes/genomes.json",
trackRegistryFile: "resources/tracks/trackRegistry.json",
// Supply a Google client id and apiKey to enable the Google file picker in the load menus. This is optional
// clientId: "...",
// apiKey: "...",
// Provide a URL shorterner function or object. This is optional. If not supplied
// sharable URLs will not be shortened .
urlShortener: {
provider: "tinyURL"
},
// The igv.js configuration object. To customize see the [igv.js wiki](https://github.com/igvteam/igv.js/wiki/Browser-Creation). If
// using a custom reference genome use a ```reference``` object in lieu of the ```genome``` property as described in the igv.js wiki.
igvConfig:
{
queryParametersSupported: true,
showChromosomeWidget: true,
genome: "hg38",
showSVGButton: false,
tracks: [
// TODO -- add default tracks here. See github.com/igvteam/igv.js/wiki for details
// {
// name: "CTCF - string url",
// type: "wig",
// format: "bigwig",
// url: "https://www.encodeproject.org/files/ENCFF563PAW/@@download/ENCFF563PAW.bigWig"
// }
]
}
}The Tracks pulldown menu is different depending on the currently selected reference genome. The set of items presented in the menu are defined in the file specified by the trackRegistryFile property in igvwebConfig.js by associating lists of track configuration files with genome IDs.
The example track registry file below will result in two items for the Tracks menu when the hg19 genome is seleted, and a single menu item for hg38. Note: make sure to include the comma after every item in the list, except the last one.
{
"hg19" : [
"resources/tracks/hg19_annotations.json",
"resources/tracks/hg19_my_platinum_genomes.json"
],
"hg38" : [
"resources/tracks/hg38_annotations.json"
]
}Each .json track configuration file associated with a genome ID corresponds to one menu item presented in the Tracks pulldown menu. The configuration file defines:
- The label of the item in the menu.
- An optional description.
- The set of tracks presented for selection when the user clicks on the menu item. The tracks can be presented as
- a simple list of track names (default),
- or, an annotated table of tracks, defined inline in the JSON or in a separate file.
1. Simple list of track names. The example below defines a menu item labeled "Annotations" that presents a list of just one track, a BED file of Gencode gene annotations. The details of the track are defined by a track configuration object as documented in the igv.js wiki.
{
"label": "Annotations",
"description": "Annotations - source <a href=http://hgdownload.soe.ucsc.edu/downloads.html target=_blank>UCSC Genome Browser</a>",
"tracks": [
{
"type": "bed",
"url": "https://s3.amazonaws.com/igv.org.test/data/gencode.v18.collapsed.bed",
"indexURL": "https://s3.amazonaws.com/igv.org.test/data/gencode.v18.collapsed.bed.idx",
"name": "Gencode V18"
}
]
} 2. Annotated table of tracks. Setting a type property in the track configuration file to "custom-data-modal" specifies that a custom annotated table of tracks will be used. Each row in the table corresponds to a track, and columns represent attributes of the track. The resulting table can be sorted and filtered interactively by the user based on column values. Selecting a row in the table will load the corresponding track.
The data property in the configuration file defines the attribute values for each row, and also the details of the track to be loaded when the row is selected. The track details are used to create a track configuration object as documented in the igv.js wiki. The data property can either point to an external tab- or comma-delimited file, or contain the data inline as JSON, as shown in the two examples below. The columns property in the configuration file defines which track attributes are displayed in the table. An optional columnDefs property can be used to rename the column headers displayed in the table.
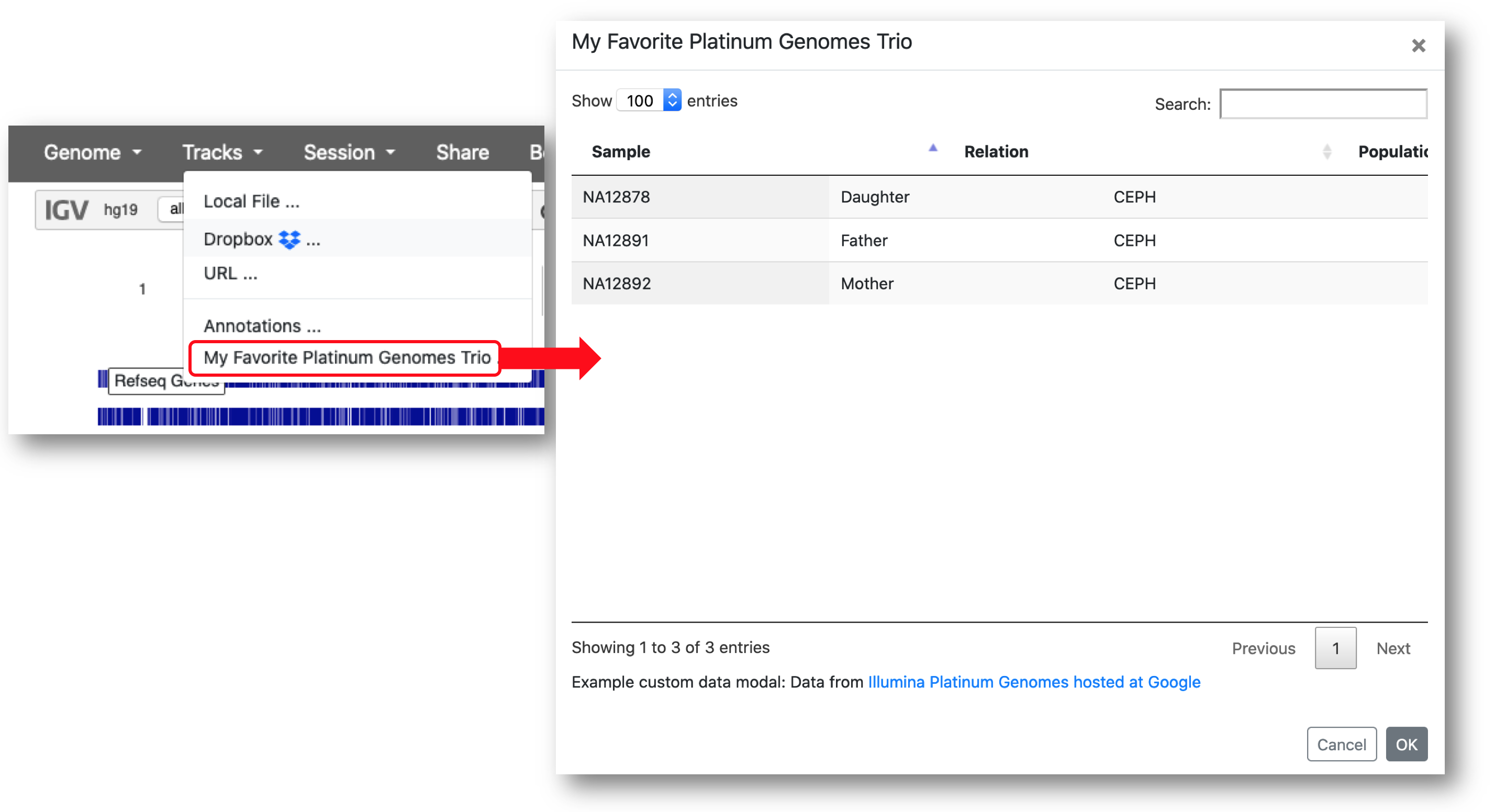
The following example defines a menu item labeled "My Favorite Platinum Genomes Trio", sets the type property to "custom-data-modal", and defines the data for the table inline as JSON. The resulting table has 3 tracks and displays 3 attributes for each. The property columnDefs is used to set the displayed column heading for the track attribute "name" to "Sample".
{
"label": "My Favorite Platinum Genomes Trio",
"type": "custom-data-modal",
"description": "Example custom data modal: Data from <a href=https://cloud.google.com/genomics/docs/public-datasets/illumina-platinum-genomes target=_blank>Illumina Platinum Genomes hosted at Google</a>",
"columns":
[
"name",
"Relation",
"Population"
],
"columnDefs":
{
"name":
{
"title": "Sample"
}
},
"data":
[
{
"Relation": "Daughter",
"Population": "CEPH",
"sourceType": "gcs",
"type": "alignment",
"url": "gs://genomics-public-data/platinum-genomes/bam/NA12878_S1.bam",
"indexURL": "gs://genomics-public-data/platinum-genomes/bam/NA12878_S1.bam.bai",
"name": "NA12878"
},
{
"Relation": "Father",
"Population": "CEPH",
"sourceType": "gcs",
"type": "alignment",
"url": "gs://genomics-public-data/platinum-genomes/bam/NA12891_S1.bam",
"indexURL": "gs://genomics-public-data/platinum-genomes/bam/NA12891_S1.bam.bai",
"name": "NA12891"
},
{
"Relation": "Mother",
"Population": "CEPH",
"sourceType": "gcs",
"type": "alignment",
"url": "gs://genomics-public-data/platinum-genomes/bam/NA12892_S1.bam",
"indexURL": "gs://genomics-public-data/platinum-genomes/bam/NA12892_S1.bam.bai",
"name": "NA12892"
}
]
}The following example shows the track configuration file for the same menu item and table as the example above, but in this case the track details are in a file located in the resources/tracks directory, as specified by the data property. Alternatively, the data property can specify a URL to the file.
{
"label": "My Favorite Platinum Genomes Trio",
"type": "custom-data-modal",
"description": "Example custom data modal: Data from <a href=https://cloud.google.com/genomics/docs/public-datasets/illumina-platinum-genomes target=_blank>Illumina Platinum Genomes hosted at Google</a>",
"columns":
[
"name",
"Relation",
"Population"
],
"columnDefs":
{
"name":
{
"title": "Sample"
}
},
"data": "resources/tracks/my_custom_track_table.csv"
}The data file can be tab- or comma-separated, specified by setting a delimeter property to "," or "\t". If the property is not set, the .csv or .tab filename extensions specify the file type. The file in this example is a .csv file. It has the same information as the JSON example above, but formatted as follows:
Relation,Population,sourceType,type,url,indexURL,name
Daughter,CEPH,gcs,alignment,gs://genomics-public-data/platinum-genomes/bam/NA12878_S1.bam,gs://genomics-public-data/platinum-genomes/bam/NA12878_S1.bam.bai,NA12878
Father,CEPH,gcs,alignment,gs://genomics-public-data/platinum-genomes/bam/NA12891_S1.bam,gs://genomics-public-data/platinum-genomes/bam/NA12891_S1.bam.bai,NA12891
Mother,CEPH,gcs,alignment,gs://genomics-public-data/platinum-genomes/bam/NA12892_S1.bam,gs://genomics-public-data/platinum-genomes/bam/NA12892_S1.bam.bai,NA12892
IGV-Web uses igv.js, a JavasScript client. To host your own track data, note that the hosting server must support CORS access and Range requests. See https://github.com/igvteam/igv.js/wiki/Data-Server-Requirements for more details.
The IGV-Web app is MIT licensed.