gmmDenoise is a set of functions for filtering erroneous sequences or amplicon sequence variants (ASVs) in eDNA metabarcoding data, based on Gaussian mixture modeling (GMM).
# install.packages("devtools")
devtools::install_github("YSKoseki/gmmDenoise")This is an example of how gmmDenoise works for the filtering of ASVs.
library(gmmDenoise)# Data: a vector of 1,217 ASV read counts, named with assigned taxonomic names
# and [ID numbers]
data(mifish)
head(mifish, n = 10)
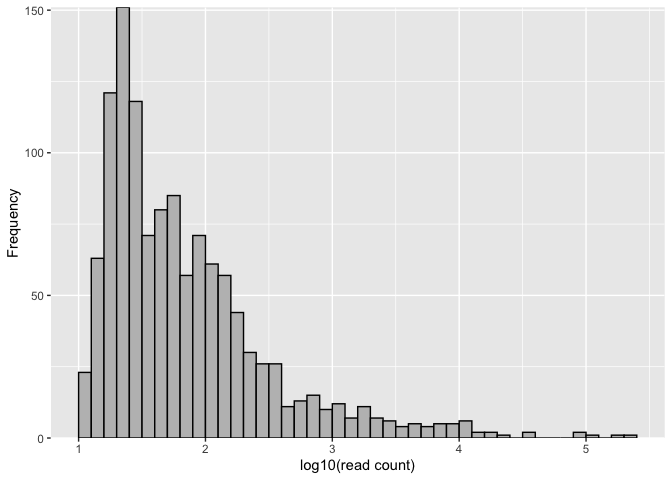
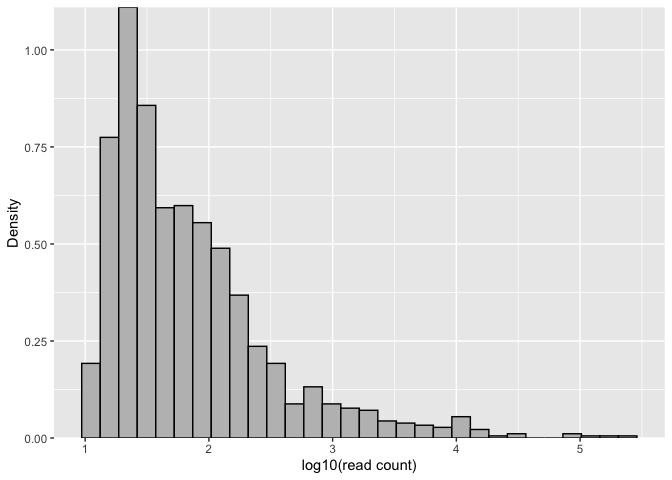
# Plot histogram for visual inspection of ASV read count distribution
asvhist(mifish)asvhist(mifish, type = "density", nbins = 30, xlim = c(1, 6))# Cross-validation analysis for selecting the number of components of Gaussian
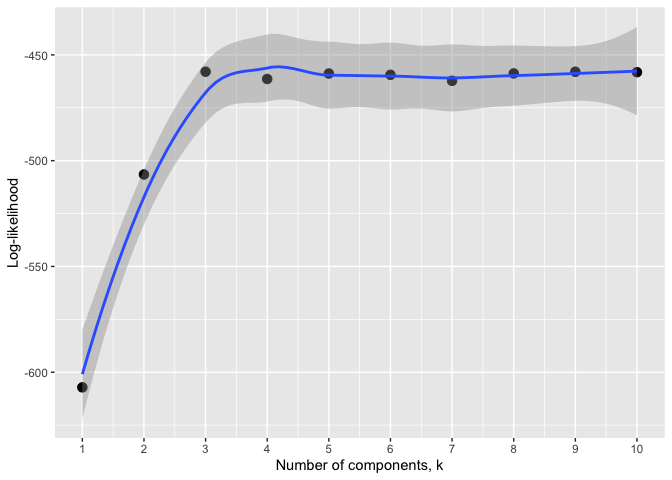
# mixture model
logmf <- log10(mifish)
set.seed(101)
cv <- gmmcv(logmf, epsilon = 1e-03)
autoplot(cv) # equivalent to `autoplot.gmmcv(cv)`# An alternative approach for the number of mixture components: Sequential
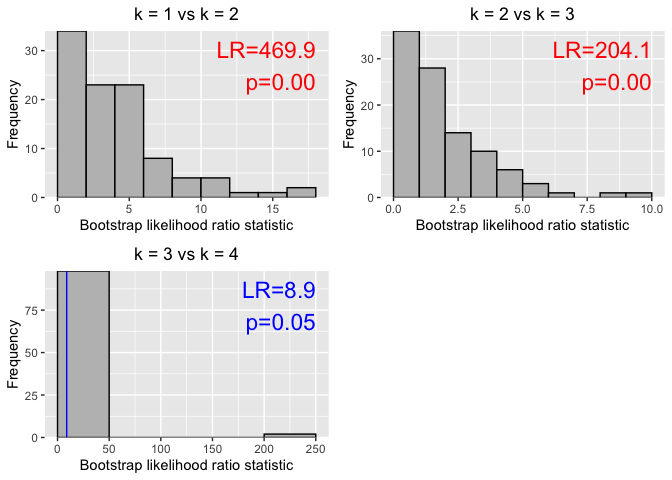
# parametric bootstrap tests
set.seed(101)
# May take some time
bs <- gmmbs(logmf, B = 100, epsilon = 1e-03)
p <- autoplot(bs) # equivalent to `p <- autoplot.gmmbs(bs)`
library(cowplot)
plot_grid(plotlist = p, ncol = 2)summary(bs)
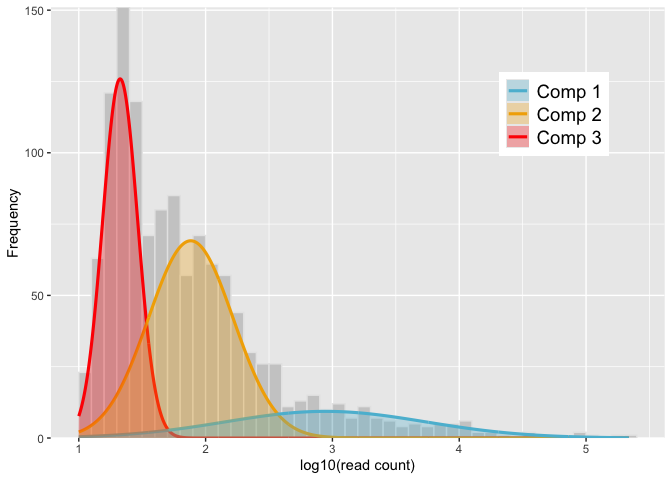
# Fit 3-component Gaussian mixture model and display a graphical representation
# of the output
set.seed(101)
mod <- gmmem(logmf, k = 3)
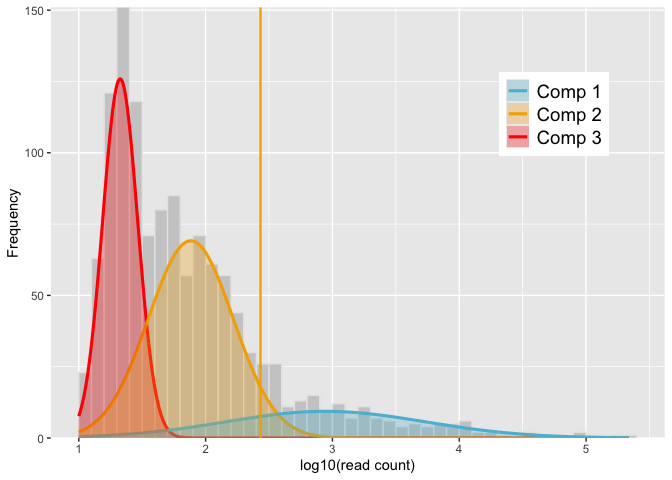
autoplot(mod) # equivalent to `autoplot.gmmem(mod)`thresh <- quantile(mod, comp = 2)
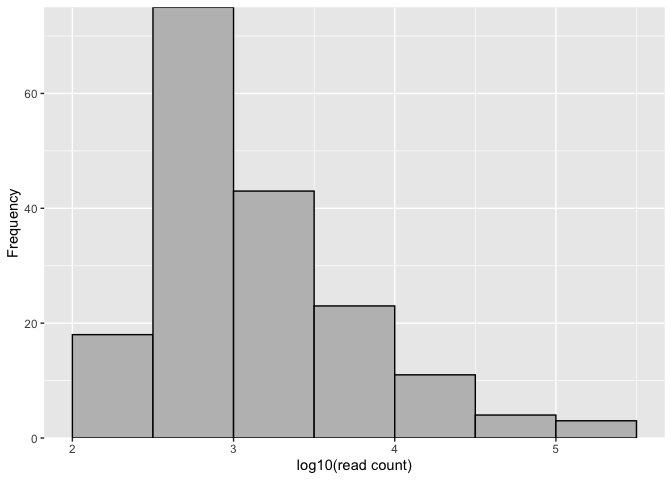
autoplot(mod, vline = c(NA, thresh, NA))# Filter ASVs with the threshold value
logmf2 <- logmf[which(logmf > thresh)]
mifish2 <- mifish[which(logmf > thresh)]
asvhist(mifish2)