Scalable De Novo Isoform Discovery
IsoSeq3 contains the newest tools to identify transcripts in PacBio single-molecule sequencing data. Starting in SMRT Link v6.0.0, those tools power the IsoSeq3 GUI-based analysis application. A composable workflow of existing tools and algorithms, combined with a new clustering technique, allows to process the ever-increasing yield of PacBio machines with similar performance to IsoSeq1 and IsoSeq2.
The latest pre-release, developers-only linux binaries can be found under releases or installed via bioconda:
conda install isoseq3
These binaries are not ISO compliant. For research only. Not for use in diagnostics procedures.
Official support is only provided for official and stable SMRT Link builds provided by PacBio.
Unofficial support for binary pre-releases is provided via github issues, not via mail to developers.
Binaries require SSE4.1 CPU support; CPUs after 2008 (Penryn) include it.
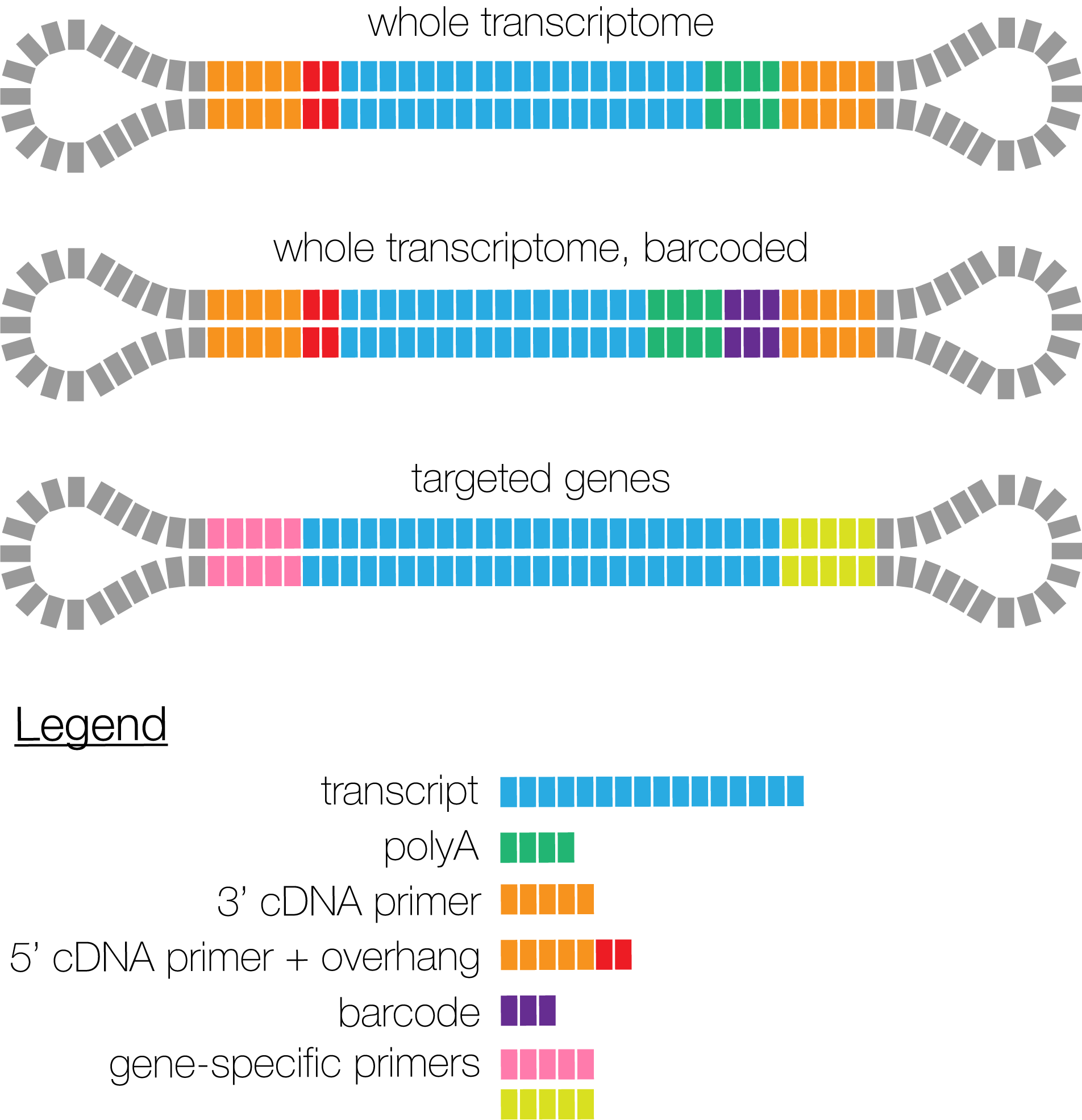
PacBio supports three different SMRTbell designs for IsoSeq libraries. In all designs, transcripts are labelled with asymmetric primers, whereas a polyA tail is optional. For whole-genome libraries, barcodes can be attached to the 3' primer.
For each cell, the <movie>.subreads.bam and <movie>.subreads.bam.pbi
are needed for processing.
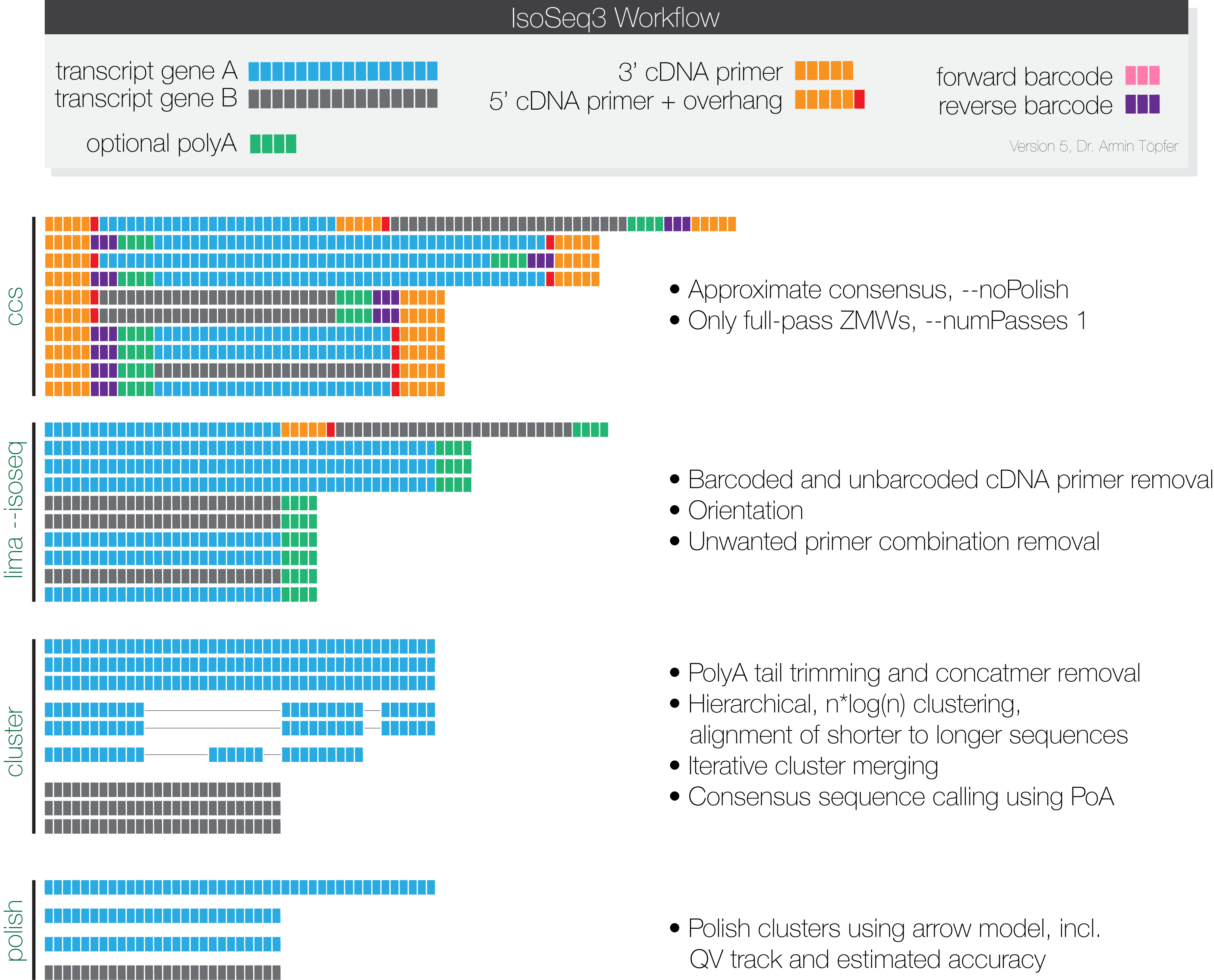
Each sequencing run is processed by ccs to generate one representative consensus sequence for each ZMW. Only ZMWs with at least one full pass, meaning at least each primer has been seen once, are used for the subsequent analysis. Furthermore, polishing is not necessary in this step and is deactivated.
ccs movie.subreads.bam ccs.bam --no-polish --num-passes 1
Removal of primers and identification of barcodes is performed using lima,
which offers a specialized --isoseq mode.
Even in the case that your sample is not barcoded, primer removal is performed
by lima.
More information about how to name input primer(+barcode)
sequences in this FAQ.
lima ccs.bam barcoded_primers.fasta demux.ccs.bam --isoseq --no-pbi
The following is the primer.fasta for the Clontech SMARTer cDNA library prep, which is the officially recommended protocol:
>primer_5p
AAGCAGTGGTATCAACGCAGAGTACATGGG
>primer_3p
GTACTCTGCGTTGATACCACTGCTT
The following are examples for barcoded samples using a 16bp barcode followed by Clontech primer:
>primer_5p
AAGCAGTGGTATCAACGCAGAGTACATGGGG
>brain_3p
CGCACTCTGATATGTGGTACTCTGCGTTGATACCACTGCTT
>liver_3p
CTCACAGTCTGTGTGTGTACTCTGCGTTGATACCACTGCTT
Lima will remove unwanted combinations and orient sequences according to the asymmetry of the primers.
From here on, execute the following steps for each output BAM file.
IsoSeq3 wraps all tools into one fat binary.
$ isoseq3
isoseq3 - De Novo Transcript Reconstruction
Tools:
cluster - Cluster CCS reads to transcripts
polish - Polish the clustering output
summarize - Create a barcode overview CSV file
Examples:
isoseq3 cluster movie.consensusreadset.xml unpolished.bam
isoseq3 polish unpolished.bam movie.subreadset.xml polished.bam
isoseq3 summarize polished.bam summary.csv
Compared to previous IsoSeq approaches, IsoSeq3 performs a single clustering technique. Due to the nature of the algorithm, it can't be efficiently chunked without creating IO bottlenecks; it is advised to give this step as many cores as possible. The individual steps of cluster are as following:
- Trimming of polyA tails
- Rapid concatmer identification and removal
- Clustering using hierarchical n*log(n) alignment and iterative cluster merging
- Unpolished POA sequence generation
The input file for cluster is one demultiplexed CCS file:
<demux.ccs.bam>or<demux.ccs.consensusreadset.xml>
The following output files of cluster contain unpolished isoforms:
<prefix>.bam<prefix>.flnc.bam<prefix>.fasta<prefix>.bam.pbi<- Only generated with--pbi<prefix>.transcriptset.xml<- Only relevant for pbsmrtpipe<prefix>.consensusreadset.xml<- Only relevant for pbsmrtpipe
Example invocation:
isoseq3 cluster demux.P5--P3.bam unpolished.bam --verbose
Polishing via the tool polish is an optional step, but highly recommended.
The algorithm behind polish is the arrow model that also used for CCS
generation and polishing of de-novo assemblies. This step can be massively
parallelized by splitting the unpolished.bam file. Split BAM files can be
generated by cluster.
The input files for polish are:
<unpolished>.bamor<unpolished>.transcriptset.xml<movie_name>.subreads.bamor<movie_name>.subreadset.xml
The following output files of polish contain polished isoforms:
<prefix>.bam<prefix>.bam.pbi<- Only generated with--pbi<prefix>.transcriptset.xml<prefix>.hq.fasta.gzwith predicted accuracy ≥ 0.99<prefix>.lq.fasta.gzwith predicted accuracy < 0.99<prefix>.hq.fastq.gzwith predicted accuracy ≥ 0.99<prefix>.lq.fastq.gzwith predicted accuracy < 0.99
Example invocation:
isoseq3 polish unpolished.bam m54020_171110_2301211.subreads.bam polished.bam
- ccs: Get it from the official SMRT Link or compile your own from unanimity
- lima: Pre-compiled binary from barcoding
- isoseq3: Pre-compiled binaries from releases
Add the directory containing the binaries to PATH:
export PATH=$PATH:<path_to_binaries>
This is an example of an end-to-end cmd-line-only workflow to get from subreads to polished isoforms; timings are system dependent:
$ wget https://downloads.pacbcloud.com/public/dataset/RC0_1cell_2017/m54086_170204_081430.subreads.bam
$ wget https://downloads.pacbcloud.com/public/dataset/RC0_1cell_2017/m54086_170204_081430.subreads.bam.pbi
$ ccs --version
ccs 3.0.0 (commit f9f505c)
$ time ccs m54086_170204_081430.subreads.bam m54086_170204_081430.ccs.bam \
--noPolish --minPasses 1
real 50m43.090s
user 3531m35.620s
sys 24m36.884s
$ cat primers.fasta
>primer_5p
AAGCAGTGGTATCAACGCAGAGTACATGGGG
>primer_3p
AAGCAGTGGTATCAACGCAGAGTAC
$ lima --version
lima 1.6.1 (commit v1.6.1-1-g77bd658)
$ time lima m54086_170204_081430.ccs.bam primers.fasta demux.bam \
--isoseq --no-pbi --dump-clips
real 0m6.543s
user 0m51.170s
$ ls demux*
demux.json demux.lima.counts demux.lima.report demux.lima.summary demux.primer_5p--primer_3p.bam demux.primer_5p--primer_3p.subreadset.xml
$ time isoseq3 cluster demux.primer_5p--primer_3p.bam unpolished.bam --verbose
Read BAM : (200740) 8s 313ms
India : (197869) 9s 204ms
Save flnc file : 35s 366ms
Convert to reads : 36s 967ms
Sort Reads : 69ms 756us
Aligning Linear : 42s 620ms
Read to clusters : 7s 506ms
Aligning Linear : 37s 595ms
Merge by mapping : 37s 645ms
Consensus : 1m 47s
Merge by mapping : 8s 861ms
Consensus : 12s 633ms
Write output : 3s 265ms
Complete run time : 5m 12s
real 5m12.888s
user 58m35.243s
$ ls unpolished*
unpolished.bam unpolished.bam.pbi unpolished.cluster unpolished.fasta unpolished.flnc.bam unpolished.flnc.bam.pbi unpolished.flnc.consensusreadset.xml unpolished.transcriptset.xml
$ time isoseq3 polish unpolished.bam m54086_170204_081430.subreads.bam polished.bam --verbose
14561
real 60m37.564s
user 2832m8.382s
$ ls polished*
polished.bam polished.bam.pbi polished.hq.fasta.gz polished.hq.fastq.gz polished.lq.fasta.gz polished.lq.fastq.gz polished.transcriptset.xml
If you have multiple cells, you should run --split-bam in the cluster step which will produce chunked cluster results. Each chunked cluster result can be run as a parallel polish job and merged at the end. The following example splits into 24 chunks. sample.subreadset.xml is the dataset containing all the input cells. The isoseq3 polish jobs can be run in parallel.
$ isoseq3 cluster demux.primer_5p--primer_3p.bam unpolished.bam --split-bam 24
$ isoseq3 polish unpolished.0.bam sample.subreadset.xml polished.0.bam
$ isoseq3 polish unpolished.1.bam sample.subreadset.xml polished.1.bam
$ ...
The ever-increasing throughput of the Sequel system gave rise to the need for a scalable software solution that can handle millions of CCS reads, while maintaining sensitivity and accuracy. Internal benchmarks have shown that IsoSeq3 is orders of magnitude faster than currently employed solutions and SQANTI attributes IsoSeq3 a higher number of perfectly annotated isoforms. Additional benefit, single linux binary that requires no dependencies.
Even though we also observe fewer polished transcripts with IsoSeq3, the overall quality is much higher. Most of the low-quality transcripts are lost in the demultiplexing step. Isoseq1/2 classify is too relaxed and is not filtering junk molecules to a satifactory level. In fact, lima calls are spot on and effectively removes most molecules that are wrongly tagged, as in two 5' or two 3' primers. Only a proper 5' and 3' primer pair allows to identify a full-length transcript and its orientation.
Classify has been replaced with PacBio's standard demultiplexing tool lima. Lima does not remove polyA tails, nor detects concatmers. See the next Q.
One of the early outputs of the cluster step is a *.flnc.bam file. Feel
free to abort after this file has been written.
There is no ETA feature. Depending on the sample type, whole transcriptome or targeted amplification, run time varies. The same number of reads from a whole transcriptome sample can finish clustering in minutes, whereas a single gene amplification of 10kb transcripts can take a couple of hours.
In contrast to its predecessors, IsoSeq3 does not rely on NP-hard clique
finding, but uses a hierarchical alignment strategy with O(N*log(N)).
Recent advances in rapid alignment of long reads make this this approach
feasible.
Cluster uses up to 10 CCS reads to generate the unpolished cluster consensus.
Polish uses up to 60 subreads to polish the cluster consensus.
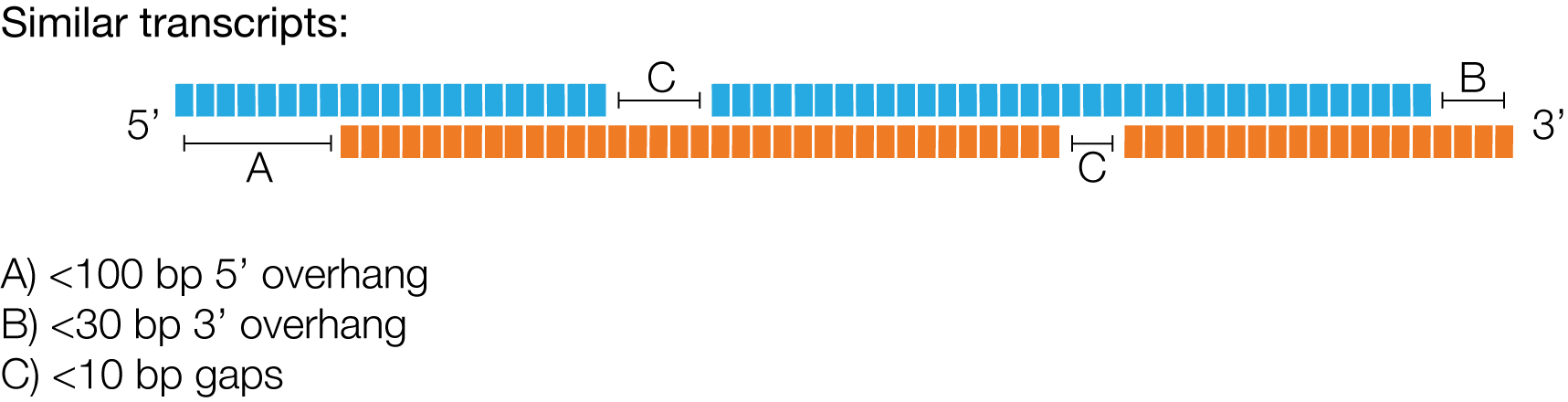
IsoSeq3 deems two reads to stem from the same transcript, if they meet following criteria:
There is no upper limit on the number of gaps.
Following BAM tags are being used:
ibBarcode summary: triplets delimited by semicolons, each triplet contains two barcode indices and the ZMW counts, delimited by comma. Example:0,1,20;0,3,5imNumber of ZMWs associated with this isoformisZMW names associated with this isoformizMaximum number of subreads used for polishingrqPredicted accuracy for polished isoform
Quality values are capped at 93.
THIS WEBSITE AND CONTENT AND ALL SITE-RELATED SERVICES, INCLUDING ANY DATA, ARE PROVIDED "AS IS," WITH ALL FAULTS, WITH NO REPRESENTATIONS OR WARRANTIES OF ANY KIND, EITHER EXPRESS OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, ANY WARRANTIES OF MERCHANTABILITY, SATISFACTORY QUALITY, NON-INFRINGEMENT OR FITNESS FOR A PARTICULAR PURPOSE. YOU ASSUME TOTAL RESPONSIBILITY AND RISK FOR YOUR USE OF THIS SITE, ALL SITE-RELATED SERVICES, AND ANY THIRD PARTY WEBSITES OR APPLICATIONS. NO ORAL OR WRITTEN INFORMATION OR ADVICE SHALL CREATE A WARRANTY OF ANY KIND. ANY REFERENCES TO SPECIFIC PRODUCTS OR SERVICES ON THE WEBSITES DO NOT CONSTITUTE OR IMPLY A RECOMMENDATION OR ENDORSEMENT BY PACIFIC BIOSCIENCES.