Table of Contents
- VCF files
- Installation
- Usage
- Viewing a BCF file
- Comparing output types
- BCF to VCF
- Filtering for different types of mutations
- VCF to PED
- Extracting INFO field/s
- Filtering VCF on the FILTER column
- Filtering VCF file using the INFO field/s
- Summarise SNPs and INDELs per sample
- Summarise genotypes in a VCF file
- Check whether the REF sequence is correct
- Random subset of variants
- Subset variants within a specific genomic region
- Subset a single sample from a multi-sample VCF file
- Merging VCF files
- Creating a test file
- Using GATK for calling variants
- Comparing VCF files
- Visualisation
- Useful links
Created by gh-md-toc
VCF files
Natural selection relies on three conditions:
- There must be genetic variation among species
- The genetic variation must be heritable
- The genetic variation results in differing fitness
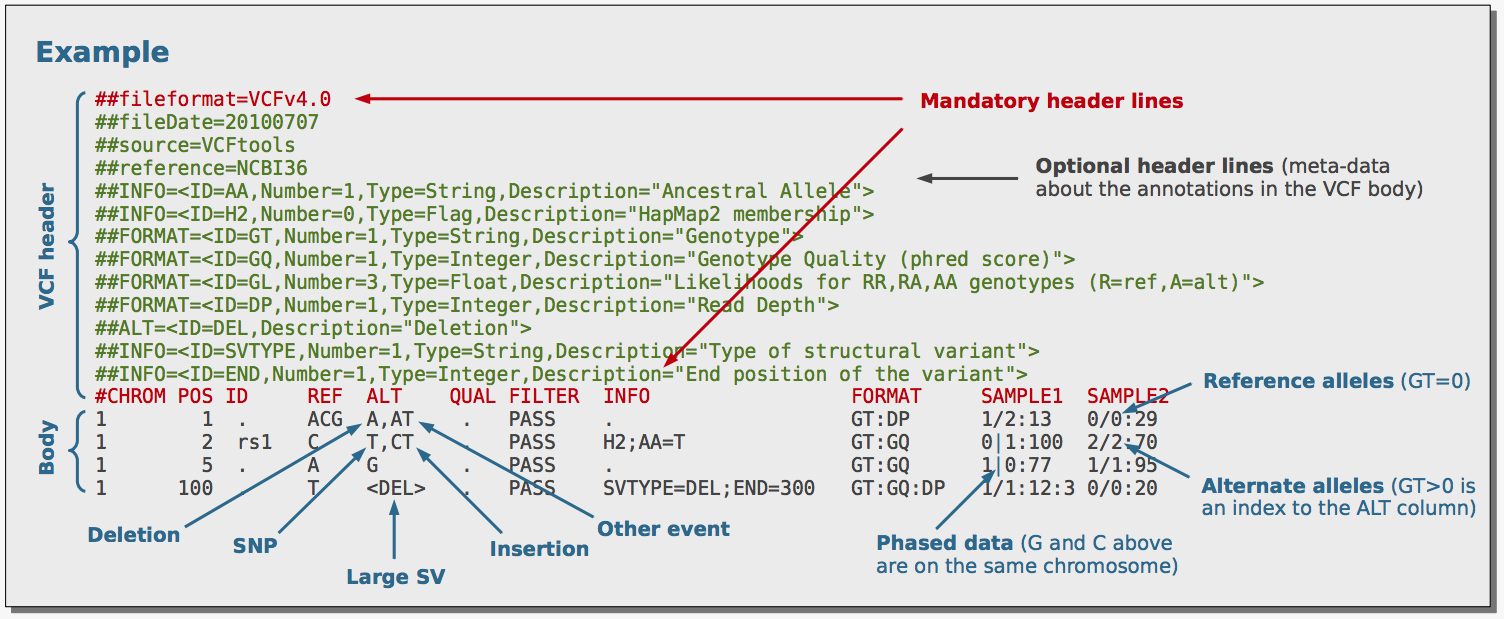
The de facto file format for representing genetic variation is the Variant Call Format (VCF). A good starting point for learning about the VCF is this poster; the image below was taken from the poster. The binary equivalent of a VCF file is a BCF file, akin to the SAM and BAM formats. BCFtools is used to view and manipulate VCF/BCF files. I have included an example BCF file (aln_consensus.bcf) in this repository to demonstrate the various utilities of BCFtools. If you are interested in how this file was generated refer to Creating a test file.
Installation
I recommend using Conda, in particular Miniconda, for installing VCFtools and BCFtools. I have created environment.yml which can be used to install all the necessary tools, used in the examples, into a new environment.
# create and install necessary programs
conda env create -f environment.yml
# activate the environment
source activate learning_vcf
# deactivate the environment you finish
source deactivate
# remove environment
# conda env remove --name learning_vcfOtherwise you can compile your own version and install BCFtools into the current directory; change --prefix if you want to install elsewhere.
mkdir bin
cd bin
wget -c https://github.com/samtools/bcftools/releases/download/1.6/bcftools-1.6.tar.bz2
tar xjf bcftools-1.6.tar.bz2
cd bcftools-1.6
./configure --prefix=`pwd`
makeInstall VCFtools.
git clone https://github.com/vcftools/vcftools.git
cd vcftools
autogen.sh
./configure --prefix=`pwd`
make
make installUsage
Typing bcftools without any parameters will output the usage and the subcommands.
bcftools
Program: bcftools (Tools for variant calling and manipulating VCFs and BCFs)
Version: 1.9 (using htslib 1.9)
Usage: bcftools [--version|--version-only] [--help] <command> <argument>
Commands:
-- Indexing
index index VCF/BCF files
-- VCF/BCF manipulation
annotate annotate and edit VCF/BCF files
concat concatenate VCF/BCF files from the same set of samples
convert convert VCF/BCF files to different formats and back
isec intersections of VCF/BCF files
merge merge VCF/BCF files files from non-overlapping sample sets
norm left-align and normalize indels
plugin user-defined plugins
query transform VCF/BCF into user-defined formats
reheader modify VCF/BCF header, change sample names
sort sort VCF/BCF file
view VCF/BCF conversion, view, subset and filter VCF/BCF files
-- VCF/BCF analysis
call SNP/indel calling
consensus create consensus sequence by applying VCF variants
cnv HMM CNV calling
csq call variation consequences
filter filter VCF/BCF files using fixed thresholds
gtcheck check sample concordance, detect sample swaps and contamination
mpileup multi-way pileup producing genotype likelihoods
roh identify runs of autozygosity (HMM)
stats produce VCF/BCF stats
Most commands accept VCF, bgzipped VCF, and BCF with the file type detected
automatically even when streaming from a pipe. Indexed VCF and BCF will work
in all situations. Un-indexed VCF and BCF and streams will work in most but
not all situations.Viewing a BCF file
I have included an example VCF file in the eg folder of this repository. Use bcftools view to view a VCF, bgzipped VCF, and BCF file.
bcftools view eg/Pfeiffer.vcf | grep -v "^#" | head -5
1 866511 rs60722469 C CCCCT 258.62 PASS AC=2;AF=1;AN=2;DB;DP=11;FS=0;HRun=0;HaplotypeScore=41.3338;MQ0=0;MQ=61.94;QD=23.51;set=variant GT:AD:DP:GQ:PL 1/1:6,5:11:14.79:300,15,0
1 879317 rs7523549 C T 150.77 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=1.455;DB;DP=21;Dels=0;FS=1.984;HRun=0;HaplotypeScore=0;MQ0=0;MQ=60;MQRankSum=-0.037;QD=7.18;ReadPosRankSum=0.112;set=variant2 GT:AD:DP:GQ:PL 0/1:14,7:21:99:181,0,367
1 879482 . G C 484.52 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=1.934;DP=48;Dels=0;FS=4.452;HRun=0;HaplotypeScore=0.5784;MQ0=0;MQ=59.13;MQRankSum=-0.24;QD=10.09;ReadPosRankSum=1.537;set=variant2 GT:AD:DP:GQ:PL 0/1:28,20:48:99:515,0,794
1 880390 rs3748593 C A 288.44 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=-4.517;DB;DP=29;Dels=0;FS=1.485;HRun=0;HaplotypeScore=0;MQ0=0;MQ=56.93;MQRankSum=-0.065;QD=9.95;ReadPosRankSum=0.196;set=variant2 GT:AD:DP:GQ:PL 0/1:14,15:29:99:318,0,399
1 881627 rs2272757 G A 486.24 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=0.199;DB;DP=33;Dels=0;FS=0;HRun=1;HaplotypeScore=1.8893;MQ0=0;MQ=60;MQRankSum=0.777;QD=14.73;ReadPosRankSum=-0.669;set=variant2 GT:AD:DP:GQ:PL 0/1:15,18:33:99:516,0,420Comparing output types
I'll use the 1000 Genomes Project WGS VCF file. There are four output types: compressed BCF (b), uncompressed BCF (u), compressed VCF (z), and uncompressed VCF (v).
# download VCF file
wget -c ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ALL.wgs.phase3_shapeit2_mvncall_integrated_v5b.20130502.sites.vcf.gz
# size of file
ls -lah ALL.wgs.phase3_shapeit2_mvncall_integrated_v5b.20130502.sites.vcf.gz
-rw-r--r-- 1 dtang dtang 1.9G Oct 12 2015 ALL.wgs.phase3_shapeit2_mvncall_integrated_v5b.20130502.sites.vcf.gz
# extract file
time gunzip -c ALL.wgs.phase3_shapeit2_mvncall_integrated_v5b.20130502.sites.vcf.gz > uncompressed.vcf
real 1m22.484s
user 0m56.220s
sys 0m25.808sHow long does it take to create the different output types?
# how long for compressed VCF?
time bcftools convert -o compressed.vcf.gz -O z uncompressed.vcf
real 11m30.628s
user 11m8.632s
sys 0m19.724s
# how long for uncompressed BCF?
time bcftools convert -o uncompressed.bcf -O u uncompressed.vcf
real 3m3.253s
user 2m41.240s
sys 0m20.040s
# how long for compressed BCF?
time bcftools convert -o compressed.bcf -O b uncompressed.vcf
real 6m50.487s
user 6m37.076s
sys 0m12.764sWhat are the file sizes?
ls -lah uncompressed.vcf
-rw-r--r-- 1 dtang dtang 12G Feb 17 16:55 uncompressed.vcf
ls -lah uncompressed.bcf
-rw-r--r-- 1 dtang dtang 9.9G Feb 17 17:14 uncompressed.bcf
ls -lah compressed.bcf
-rw-r--r-- 1 dtang dtang 2.0G Feb 17 17:22 compressed.bcf
ls -lah compressed.vcf.gz
-rw-r--r-- 1 dtang dtang 1.9G Feb 17 17:09 compressed.vcf.gzHow long to read each file?
time cat uncompressed.vcf | grep -v "^#" | wc -l
84801880
real 0m26.856s
user 0m7.696s
sys 0m41.996s
time bcftools view uncompressed.bcf | grep -v "^#" | wc -l
84801880
real 3m32.467s
user 3m31.268s
sys 0m30.992s
time bcftools view compressed.vcf.gz | grep -v "^#" | wc -l
84801880
real 6m58.366s
user 6m49.544s
sys 0m32.448s
time gunzip -c compressed.vcf.gz | grep -v "^#" | wc -l
84801880
real 1m3.143s
user 1m3.488s
sys 0m15.816s
time bcftools view compressed.bcf | grep -v "^#" | wc -l
84801880
real 4m1.538s
user 4m4.188s
sys 0m27.620sSeems like using compressed VCF is the best choice (smallest size and gunzip is much faster).
BCF to VCF
Use the convert subcommand.
# -O, --output-type <b|u|z|v> b: compressed BCF, u: uncompressed BCF, z: compressed VCF, v: uncompressed VCF [v]
# -o, --output <file> output file name [stdout]
bcftools convert -O v -o aln_consensus.vcf aln_consensus.bcf
# we can use cat to view the file
cat aln_consensus.vcf | grep -v "^#" | head
1000000 58 . AT A 77.4563 . INDEL;IDV=57;IMF=1;DP=57;VDB=1.20228e-08;SGB=-0.693136;MQ0F=0;AF1=1;AC1=2;DP4=0,0,35,0;MQ=60;FQ=-139.526 GT:PL 1/1:118,105,0
1000000 68 . CTTTT CTTT 70.4562 . INDEL;IDV=68;IMF=1;DP=68;VDB=7.54492e-06;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,51,0;MQ=60;FQ=-188.527 GT:PL 1/1:111,154,0
1000000 225 . CTT CT 169.457 . INDEL;IDV=78;IMF=0.928571;DP=84;VDB=0.0449154;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,79,0;MQ=60;FQ=-272.528 GT:PL 1/1:210,238,0
1000000 336 . A G 221.999 . DP=112;VDB=0.756462;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,102,0;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 378 . T C 221.999 . DP=101;VDB=0.704379;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,99,0;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 451 . AGG AGGG 214.458 . INDEL;IDV=127;IMF=0.969466;DP=131;VDB=0.0478427;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,87,42;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 915 . G GC 214.458 . INDEL;IDV=179;IMF=0.913265;DP=196;VDB=0.929034;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,89,101;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 1009 . G C 221.999 . DP=203;VDB=0.259231;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,94,101;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 1062 . ATT AT 214.458 . INDEL;IDV=187;IMF=0.958974;DP=195;VDB=0.244824;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,97,92;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 1207 . T G 221.999 . DP=177;VDB=0.628515;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,94,79;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0Filtering for different types of mutations
The view subcommand lets you select specific types of variants.
SNPs
bcftools view -v snps aln_consensus.bcf | grep -v "^#" | head
1000000 336 . A G 221.999 . DP=112;VDB=0.756462;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,102,0;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 378 . T C 221.999 . DP=101;VDB=0.704379;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,99,0;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 1009 . G C 221.999 . DP=203;VDB=0.259231;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,94,101;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 1207 . T G 221.999 . DP=177;VDB=0.628515;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,94,79;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 1281 . C A 221.999 . DP=154;VDB=0.286069;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,66,80;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 1405 . A T 221.999 . DP=203;VDB=0.0898873;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,104,89;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 1669 . G C 221.999 . DP=191;VDB=0.656207;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,108,73;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 1775 . C A 221.999 . DP=225;VDB=0.413906;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,101,115;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 2036 . T A 221.999 . DP=193;VDB=0.227246;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,83,98;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0
1000000 2180 . G C 221.999 . DP=211;VDB=0.123382;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,97,105;MQ=60;FQ=-281.989 GT:PL 1/1:255,255,0INDELs
bcftools view -v indels aln_consensus.bcf | grep -v "^#" | head
1000000 58 . AT A 77.4563 . INDEL;IDV=57;IMF=1;DP=57;VDB=1.20228e-08;SGB=-0.693136;MQ0F=0;AF1=1;AC1=2;DP4=0,0,35,0;MQ=60;FQ=-139.526 GT:PL 1/1:118,105,0
1000000 68 . CTTTT CTTT 70.4562 . INDEL;IDV=68;IMF=1;DP=68;VDB=7.54492e-06;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,51,0;MQ=60;FQ=-188.527 GT:PL 1/1:111,154,0
1000000 225 . CTT CT 169.457 . INDEL;IDV=78;IMF=0.928571;DP=84;VDB=0.0449154;SGB=-0.693147;MQ0F=0;AF1=1;AC1=2;DP4=0,0,79,0;MQ=60;FQ=-272.528 GT:PL 1/1:210,238,0
1000000 451 . AGG AGGG 214.458 . INDEL;IDV=127;IMF=0.969466;DP=131;VDB=0.0478427;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,87,42;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 915 . G GC 214.458 . INDEL;IDV=179;IMF=0.913265;DP=196;VDB=0.929034;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,89,101;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 1062 . ATT AT 214.458 . INDEL;IDV=187;IMF=0.958974;DP=195;VDB=0.244824;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,97,92;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 1278 . TA TAA 214.458 . INDEL;IDV=144;IMF=0.929032;DP=155;VDB=0.252598;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,65,80;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 1328 . AT A 129.457 . INDEL;IDV=177;IMF=0.988827;DP=179;VDB=1.83715e-25;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,36,18;MQ=60;FQ=-197.527 GT:PL 1/1:170,163,0
1000000 1380 . TA TAA 214.458 . INDEL;IDV=180;IMF=0.957447;DP=188;VDB=5.28227e-08;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,80,68;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0
1000000 1449 . GT G 214.458 . INDEL;IDV=210;IMF=0.972222;DP=216;VDB=0.783773;SGB=-0.693147;MQSB=1;MQ0F=0;AF1=1;AC1=2;DP4=0,0,101,109;MQ=60;FQ=-289.528 GT:PL 1/1:255,255,0VCF to PED
See my blog post.
Extracting INFO field/s
The VCF has various information fields; use the query subcommand to extract specific field/s.
bcftools query -f 'DP=%DP\tAF1=%AF1\tAC1=%AC1\tMQ=%MQ\n' aln_consensus.bcf | head
DP=57 AF1=1 AC1=2 MQ=60
DP=68 AF1=1 AC1=2 MQ=60
DP=84 AF1=1 AC1=2 MQ=60
DP=112 AF1=1 AC1=2 MQ=60
DP=101 AF1=1 AC1=2 MQ=60
DP=131 AF1=1 AC1=2 MQ=60
DP=196 AF1=1 AC1=2 MQ=60
DP=203 AF1=1 AC1=2 MQ=60
DP=195 AF1=1 AC1=2 MQ=60
DP=177 AF1=1 AC1=2 MQ=60Combining with the view subcommand:
bcftools view -v snps aln_consensus.bcf | bcftools query -f 'DP=%DP\tAF1=%AF1\tAC1=%AC1\tMQ=%MQ\n' - | head
DP=112 AF1=1 AC1=2 MQ=60
DP=101 AF1=1 AC1=2 MQ=60
DP=203 AF1=1 AC1=2 MQ=60
DP=177 AF1=1 AC1=2 MQ=60
DP=154 AF1=1 AC1=2 MQ=60
DP=203 AF1=1 AC1=2 MQ=60
DP=191 AF1=1 AC1=2 MQ=60
DP=225 AF1=1 AC1=2 MQ=60
DP=193 AF1=1 AC1=2 MQ=60
DP=211 AF1=1 AC1=2 MQ=60Filtering VCF on the FILTER column
Use bcftools view to keep variants that have a "PASS" in the FILTER column.
# -f, --apply-filters <list> require at least one of the listed FILTER strings (e.g. "PASS,.")
bcftools view -f PASS my.vcf > my_passed.vcfFiltering VCF file using the INFO field/s
Use vcffilter from vcflib, which is a C++ library for parsing and manipulating VCF files.
git clone --recursive https://github.com/vcflib/vcflib.git
cd vcflib
make
cd ..
# create VCF from BCF using
# bcftools convert -O v -o aln_consensus.vcf aln_consensus.bcf
# filter variants based on depth (DP)
vcflib/bin/vcffilter -f "DP > 200" aln_consensus.vcf | grep -v "^#" | head
1000000 1009 . G C 221.999 . AC1=2;AF1=1;DP=203;DP4=0,0,94,101;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.259231 GT:PL 1/1:255,255,0
1000000 1405 . A T 221.999 . AC1=2;AF1=1;DP=203;DP4=0,0,104,89;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.0898873 GT:PL 1/1:255,255,0
1000000 1449 . GT G 214.458 . AC1=2;AF1=1;DP=216;DP4=0,0,101,109;FQ=-289.528;IDV=210;IMF=0.972222;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.783773;INDEL GT:PL 1/1:255,255,0
1000000 1775 . C A 221.999 . AC1=2;AF1=1;DP=225;DP4=0,0,101,115;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.413906 GT:PL 1/1:255,255,0
1000000 2180 . G C 221.999 . AC1=2;AF1=1;DP=211;DP4=0,0,97,105;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.123382 GT:PL 1/1:255,255,0
1000000 2340 . TGGGGG TGGGG 214.458 . AC1=2;AF1=1;DP=201;DP4=0,0,100,98;FQ=-289.528;IDV=195;IMF=0.970149;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.064618;INDEL GT:PL 1/1:255,255,0
1000000 2717 . CAAAA CAAA 214.458 . AC1=2;AF1=1;DP=211;DP4=0,0,99,105;FQ=-289.528;IDV=202;IMF=0.957346;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.251202;INDEL GT:PL 1/1:255,255,0
1000000 3059 . T C 221.999 . AC1=2;AF1=1;DP=206;DP4=0,0,97,100;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.697352 GT:PL 1/1:255,255,0
1000000 3114 . TC T 214.458 . AC1=2;AF1=1;DP=209;DP4=0,0,76,77;FQ=-289.528;IDV=203;IMF=0.971292;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=2.55116e-09;INDEL GT:PL 1/1:255,255,0
1000000 3148 . CGGG CGG 94.4565 . AC1=2;AF1=1;DP=211;DP4=0,0,19,18;FQ=-145.526;IDV=196;IMF=0.92891;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693141;VDB=0.603852;INDEL GT:PL 1/1:135,111,0
# filter on two INFO fields
vcflib/bin/vcffilter -f "DP > 200 & VDB > 0.5" aln_consensus.vcf | grep -v "^#" | head
1000000 1449 . GT G 214.458 . AC1=2;AF1=1;DP=216;DP4=0,0,101,109;FQ=-289.528;IDV=210;IMF=0.972222;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.783773;INDEL GT:PL 1/1:255,255,0
1000000 3059 . T C 221.999 . AC1=2;AF1=1;DP=206;DP4=0,0,97,100;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.697352 GT:PL 1/1:255,255,0
1000000 3148 . CGGG CGG 94.4565 . AC1=2;AF1=1;DP=211;DP4=0,0,19,18;FQ=-145.526;IDV=196;IMF=0.92891;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693141;VDB=0.603852;INDEL GT:PL 1/1:135,111,0
1000000 3876 . C T 221.999 . AC1=2;AF1=1;DP=223;DP4=0,0,98,112;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.839919 GT:PL 1/1:255,255,0
1000000 4079 . C T 221.999 . AC1=2;AF1=1;DP=212;DP4=0,0,92,109;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.579433 GT:PL 1/1:255,255,0
1000000 4173 . CT C 214.458 . AC1=2;AF1=1;DP=207;DP4=0,0,106,91;FQ=-289.528;IDV=197;IMF=0.951691;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.874655;INDEL GT:PL 1/1:255,255,0
1000000 4642 . C T 221.999 . AC1=2;AF1=1;DP=205;DP4=0,0,94,105;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.523597 GT:PL 1/1:255,255,0
1000000 4676 . T C 221.999 . AC1=2;AF1=1;DP=203;DP4=0,0,96,102;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.611013 GT:PL 1/1:255,255,0
1000000 4689 . T C 221.999 . AC1=2;AF1=1;DP=216;DP4=0,0,98,109;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.879521 GT:PL 1/1:255,255,0
1000000 5121 . A T 221.999 . AC1=2;AF1=1;DP=213;DP4=0,0,105,104;FQ=-281.989;MQ=60;MQ0F=0;MQSB=1;SGB=-0.693147;VDB=0.743159 GT:PL 1/1:255,255,0Summarise SNPs and INDELs per sample
Use bcftools stats with the -s - parameter. The example VCF file eg/ex.vcf has four variants across three samples (one, two, and three).
- Sample one has two SNPs (both het) and one deletion (het)
- Sample two has two SNPs (one het and one hom alt) and insertion (het)
- Sample three has one SNP (het) and one insertion (hom alt) and one deletion (hom alt)
You can confirm the numbers from the stats output.
cat eg/ex.vcf | grep -v "^#"
1 866511 rs60722469 C CCCCT 258.62 PASS AC=2;AF=1.00;AN=2;DB;DP=11;FS=0.000;HRun=0;HaplotypeScore=41.3338;MQ0=0;MQ=61.94;QD=23.51;set=variant GT:AD:DP:GQ:PL 0/0:6,5:11:14.79:300,15,0 0/1:6,5:11:14.79:300,15,0 1/1:6,5:11:14.79:300,15,0
1 884091 rs7522415 C G 65.46 PASS AC=1;AF=0.50;AN=2;BaseQRankSum=-0.259;DB;DP=12;Dels=0.00;FS=0.000;HRun=1;HaplotypeScore=0.0000;MQ0=0;MQ=53.22;MQRankSum=0.779;QD=5.45;ReadPosRankSum=2.047;set=variant2 GT:AD:DP:GQ:PL 0/1:6,6:12:95.45:95,0,123 1/1:6,6:12:95.45:95,0,123 0/0:6,6:12:95.45:95,0,123
1 897730 rs7549631 C T 225.34 PASS AC=1;AF=0.50;AN=2;BaseQRankSum=-2.218;DB;DP=21;Dels=0.00;FS=6.419;HRun=1;HaplotypeScore=1.8410;MQ0=0;MQ=58.89;MQRankSum=-0.387;QD=10.73;ReadPosRankSum=-0.880;set=variant2 GT:AD:DP:GQ:PL 0/1:11,10:21:99:255,0,348 0/1:11,10:21:99:255,0,348 0/1:11,10:21:99:255,0,348
1 1158562 rs57524763 AAC A 220.99 PASS AC=1;AF=0.50;AN=2;BaseQRankSum=2.621;DB;DP=20;FS=0.000;HRun=0;HaplotypeScore=101.7487;MQ0=0;MQ=55.80;MQRankSum=-1.910;QD=11.05;ReadPosRankSum=0.400;set=variant GT:AD:DP:GQ:PL
0/1:14,6:20:99:260,0,486 0/0:14,6:20:99:260,0,486 1/1:14,6:20:99:260,0,486
bcftools stats -s - eg/ex.vcf | grep -A 4 "Per-sample counts"
# PSC, Per-sample counts. Note that the ref/het/hom counts include only SNPs, for indels see PSI. The rest include both SNPs and indels.
# PSC [2]id [3]sample [4]nRefHom [5]nNonRefHom [6]nHets [7]nTransitions [8]nTransversions [9]nIndels [10]average depth [11]nSingletons [12]nHapRef [13]nHapAlt [14]nMissing
PSC 0 one 1 0 2 1 1 1 16.0 0 0 0 0
PSC 0 two 1 1 1 1 1 1 16.0 0 0 0 0
PSC 0 three 1 0 1 1 0 2 16.0 0 0 0 0
bcftools stats -s - eg/ex.vcf | grep -A 4 "Per-Sample Indels"
# PSI, Per-Sample Indels
# PSI [2]id [3]sample [4]in-frame [5]out-frame [6]not applicable [7]out/(in+out) ratio [8]nHets [9]nAA
PSI 0 one 0 0 0 0.00 1 0
PSI 0 two 0 0 0 0.00 1 0
PSI 0 three 0 0 0 0.00 0 2Summarise genotypes in a VCF file
Use the vcffixup tool from vcflib, which can count the allele frequencies across alleles present in each sample.
cat output.vcf | grep -v "^#" | head -1
chr21 9889293 rs28676788 G A . . . GT 0/0 ./. 0/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 0/0 1/0 1/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 1/0 1/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 0/0 0/0 0/0 0/0 0/0 0/0 ./. 0/0 0/0 0/0 0/0 1/0 1/0 0/0 0/0 ./. 0/0 0/0 ./. 1/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 0/0 0/0 0/0 1/0 1/0 1/0 0/0 0/0 1/0 1/0
vcffixup output.vcf | grep -v "^#" | head -1
chr21 9889293 rs28676788 G A 0 . AC=16;AF=0.101266;AN=158;NS=83 GT 0/0 ./. 0/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 0/0 1/0 1/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 1/0 1/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 0/0 0/0 0/0 0/0 0/0 0/0 ./. 0/0 0/0 0/0 0/0 1/0 1/0 0/0 0/0 ./. 0/0 0/0 ./. 1/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 1/0 0/0 0/0 0/0 1/0 1/0 1/0 0/0 0/0 1/0 1/0NS refers to the number of calls, i.e. the number of samples. 4 samples had no genotype, i.e. ./., therefore AN is 79*2 = 158. AC is the alternate allele count and AF is the alternate allele frequency. I asked the question of how I can summarise genotypes in a VCF file on Biostars in 2015 and ended up answering my own question 17 months later.
Using the example bgzipped VCF file I have in the plink folder.
gunzip -c plink/ex2.vcf.gz
##fileformat=VCFv4.0
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA00001 NA00002 NA00003 NA00004 NA00005 NA00006 NA00007 NA00008 NA00009 NA00010
1 10000 . C T 99 PASS DP=14 GT 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0
2 20000 . G A 99 PASS DP=14 GT 0/1 0/1 0/1 0/1 0/1 0/0 0/0 0/0 0/0 0/0
2 25000 . G A 99 PASS DP=14 GT 0/1 0/1 0/1 0/1 0/1 0/0 0/0 0/0 0/0 0/0
3 30000 . T A 99 PASS DP=11 GT 0/0 0/0 0/0 0/0 0/0 0/1 0/1 0/1 0/1 0/1
4 40000 . A G 99 PASS DP=10 GT 1/1 0/0 1/1 0/0 1/1 0/0 1/1 0/0 1/1 0/0
5 50000 . T G 99 PASS DP=13 GT 0/0 1/1 0/0 1/1 0/0 1/1 0/0 1/1 0/0 1/1
6 60000 . T C 99 PASS DP=13 GT 0/0 0/0 0/0 0/0 0/0 0/1 0/1 0/1 0/1 0/1
7 70000 . C G 99 PASS DP=9 GT 1/1 1/1 1/1 1/1 1/1 1/1 1/1 1/1 1/1 1/1
8 80000 . C G 99 PASS DP=9 GT 1/1 1/1 1/1 1/1 1/1 0/0 0/0 0/0 0/0 0/0
../vcflib/bin/vcffixup plink/ex2.vcf.gz
##fileformat=VCFv4.0
##INFO=<ID=AC,Number=A,Type=Integer,Description="Total number of alternate alleles in called genotypes">
##INFO=<ID=AF,Number=A,Type=Float,Description="Estimated allele frequency in the range (0,1]">
##INFO=<ID=NS,Number=1,Type=Integer,Description="Number of samples with data">
##INFO=<ID=AN,Number=1,Type=Integer,Description="Total number of alleles in called genotypes">
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA00001 NA00002 NA00003 NA00004 NA00005 NA00006 NA00007 NA00008 NA00009 NA00010
1 10000 . C T 99 PASS AC=0;AF=0;AN=20;DP=14;NS=10 GT 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0 0/0
2 20000 . G A 99 PASS AC=5;AF=0.25;AN=20;DP=14;NS=10 GT 0/1 0/1 0/1 0/1 0/1 0/0 0/0 0/0 0/0 0/0
2 25000 . G A 99 PASS AC=5;AF=0.25;AN=20;DP=14;NS=10 GT 0/1 0/1 0/1 0/1 0/1 0/0 0/0 0/0 0/0 0/0
3 30000 . T A 99 PASS AC=5;AF=0.25;AN=20;DP=11;NS=10 GT 0/0 0/0 0/0 0/0 0/0 0/1 0/1 0/1 0/1 0/1
4 40000 . A G 99 PASS AC=10;AF=0.5;AN=20;DP=10;NS=10 GT 1/1 0/0 1/1 0/0 1/1 0/0 1/1 0/0 1/1 0/0
5 50000 . T G 99 PASS AC=10;AF=0.5;AN=20;DP=13;NS=10 GT 0/0 1/1 0/0 1/1 0/0 1/1 0/0 1/1 0/0 1/1
6 60000 . T C 99 PASS AC=5;AF=0.25;AN=20;DP=13;NS=10 GT 0/0 0/0 0/0 0/0 0/0 0/1 0/1 0/1 0/1 0/1
7 70000 . C G 99 PASS AC=20;AF=1;AN=20;DP=9;NS=10 GT 1/1 1/1 1/1 1/1 1/1 1/1 1/1 1/1 1/1 1/1
8 80000 . C G 99 PASS AC=10;AF=0.5;AN=20;DP=9;NS=10 GT 1/1 1/1 1/1 1/1 1/1 0/0 0/0 0/0 0/0 0/0Check whether the REF sequence is correct
Use vcfcheck from vcflib.
# make another copy of the VCF file
cp aln_consensus.vcf blah.vcf
# manually change REF sequence at pos 336
# vcfcheck identifies the mismatch and reports it
vcflib/bin/vcfcheck -f test_31.fa blah.vcf
mismatched reference T should be A at 1000000:336
rm blah.vcfOtherwise you can use the simple Perl script that I wrote in the script directory. The script obtains the sequence from a fasta file based on the position reported in the VCF file and compares it to the reported reference base.
script/check_ref.pl
Usage: script/check_ref.pl <genome.fa> <infile.vcf>Random subset of variants
Use vcfrandomsample from vcflib. Below is the usage:
vcflib/bin/vcfrandomsample
usage: vcfrandomsample [options] [<vcf file>]
options:
-r, --rate RATE base sampling probability per locus
-s, --scale-by KEY scale sampling likelihood by this Float info field
-p, --random-seed N use this random seed (by default read from /dev/random)
-q, --pseudorandom-seed use a pseudorandom seed (by default read from /dev/random)
Randomly sample sites from an input VCF file, which may be provided as stdin.
Scale the sampling probability by the field specified in KEY. This may be
used to provide uniform sampling across allele frequencies, for instance.vcfrandomsample can read from STDOUT.
bcftools view aln_consensus.bcf | grep -v "^#" | wc -l
9704
# ~1%
bcftools view aln_consensus.bcf | vcflib/bin/vcfrandomsample -p 31 -r 0.01 | grep -v "^#" | wc -l
90
# ~10%
bcftools view aln_consensus.bcf | vcflib/bin/vcfrandomsample -p 31 -r 0.1 | grep -v "^#" | wc -l
948Subset variants within a specific genomic region
Use vcfintersect from vcflib by creating a BED file with your region of interest, for example where your gene is located.
vcfintersect -b my_file.bed my_file.vcf > my_subsetted_file.vcfAnother option is to use bcftools view but you can only subset one region manually.
# -t, --targets chr|chr:pos|chr:from-to|chr:from-[,...]
bcftools view -t 1:866511-882000 eg/Pfeiffer.vcf
# VCF header not shown
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT manuel
1 866511 rs60722469 C CCCCT 258.62 PASS AC=2;AF=1;AN=2;DB;DP=11;FS=0;HRun=0;HaplotypeScore=41.3338;MQ0=0;MQ=61.94;QD=23.51;set=variant GT:AD:DP:GQ:PL 1/1:6,5:11:14.79:300,15,0
1 879317 rs7523549 C T 150.77 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=1.455;DB;DP=21;Dels=0;FS=1.984;HRun=0;HaplotypeScore=0;MQ0=0;MQ=60;MQRankSum=-0.037;QD=7.18;ReadPosRankSum=0.112;set=variant2 GT:AD:DP:GQ:PL 0/1:14,7:21:99:181,0,367
1 879482 . G C 484.52 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=1.934;DP=48;Dels=0;FS=4.452;HRun=0;HaplotypeScore=0.5784;MQ0=0;MQ=59.13;MQRankSum=-0.24;QD=10.09;ReadPosRankSum=1.537;set=variant2 GT:AD:DP:GQ:PL 0/1:28,20:48:99:515,0,794
1 880390 rs3748593 C A 288.44 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=-4.517;DB;DP=29;Dels=0;FS=1.485;HRun=0;HaplotypeScore=0;MQ0=0;MQ=56.93;MQRankSum=-0.065;QD=9.95;ReadPosRankSum=0.196;set=variant2 GT:AD:DP:GQ:PL 0/1:14,15:29:99:318,0,399
1 881627 rs2272757 G A 486.24 PASS AC=1;AF=0.5;AN=2;BaseQRankSum=0.199;DB;DP=33;Dels=0;FS=0;HRun=1;HaplotypeScore=1.8893;MQ0=0;MQ=60;MQRankSum=0.777;QD=14.73;ReadPosRankSum=-0.669;set=variant2 GT:AD:DP:GQ:PL 0/1:15,18:33:99:516,0,420Subset a single sample from a multi-sample VCF file
Use GATK SelectVariants; check link out for more subsetting recipes. The -fraction also creates a random subset of variants.
# include this if you want to exclude homozygous reference
# --excludeNonVariants \
java -Xmx2g -jar GenomeAnalysisTK.jar \
-R ucsc.hg19.fasta \
-T SelectVariants \
--variant multi_sample.vcf \
-o output.vcf \
--keepOriginalAC \
-sn SAMPLE1 \
-sn SAMPLE2
# Select a sample and restrict the output VCF to a set of intervals:
java -Xmx2g -jar GenomeAnalysisTK.jar \
-R ucsc.hg19.fasta \
-T SelectVariants \
-V input.vcf \
-o output.vcf \
-L /path/to/my.interval_list \
-sn SAMPLE1 \
-sn SAMPLE2The my.interval_list file can be in several formats including the popular BED format. The GATK engine recognises the .bed extension and interprets the coordinate system accordingly.
Merging VCF files
The NHLBI Exome Sequencing Project (ESP) provides their variants in the VCF but per chromsome.
wget -c http://evs.gs.washington.edu/evs_bulk_data/ESP6500SI-V2-SSA137.GRCh38-liftover.snps_indels.vcf.tar.gz
tar -xzf ESP6500SI-V2-SSA137.GRCh38-liftover.snps_indels.vcf.tar.gz
ls -1
ESP6500SI-V2-SSA137.GRCh38-liftover.chr10.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr11.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr12.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr13.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr14.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr15.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr16.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr17.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr18.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr19.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr1.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr20.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr21.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr22.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr2.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr3.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr4.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr5.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr6.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr7.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr8.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chr9.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chrX.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.chrY.snps_indels.vcf
ESP6500SI-V2-SSA137.GRCh38-liftover.snps_indels.vcf.tar.gzWe can use bcftools merge to merge the VCF files together. The VCF files need to be compressed with bgzip and tabix indexed in order for bcftools merge to work.
# I make use of GNU parallel to speed things up
# assuming that only the ESP VCF files are in the directory
parallel bgzip ::: *.vcf
parallel tabix -p vcf ::: *.vcf.gz
# -O z for compressed VCF
bcftools merge -o ESP6500SI-V2-SSA137.all.vcf.gz -O z *.vcf.gz
# sanity check
# number of variants from the separate VCF files
gunzip -c *indels.vcf.gz | grep -v "^#" | wc -l
1986331
# number of variants in the merged VCF file
gunzip -c ESP6500SI-V2-SSA137.all.vcf.gz | grep -v "^#" | wc -l
1986331
# finally tabix index
tabix -p vcf ESP6500SI-V2-SSA137.all.vcf.gzCreating a test file
The aln_consensus.bcf file was created from a simple pipeline. Firstly a random reference sequence was generated; genetic variants are created by modifying the reference sequence, i.e. introducing mutations, into a mutated copy and sequence reads were derived from the mutated reference sequence. Lastly, the reads were mapped back to the original non-mutated reference sequence. The pipeline.groovy file contains the pipeline, which is written in Groovy and processed by Bpipe. I have a blog post that provides more information.
To create aln_consensus.bcf, simply clone this repository and type make. This will download and install all the necessary programs from online and run the pipeline.
git clone https://github.com/davetang/learning_vcf_file.git
makeAlternatively, use Conda to install all the necessary tools and use analysis/run.sh.
conda env create -f environment.yml
cd analysis
wget https://github.com/broadinstitute/gatk/releases/download/4.1.1.0/gatk-4.1.1.0.zip
unzip gatk-4.1.1.0.zip
rm gatk-4.1.1.0.zip
./run.shAdjusting parameters
All the variables are defined in pipeline.groovy, which can be adjusted.
SEED=31
REF_SIZE=1000000
REF="test_" + "$SEED" + ".fa"
REF_MUT="test_mutated.fa"
REF_MUT_LOG="test_mutated.log"
MUT_PC=0.01
//READ_NO=300000
READ_NO=1000000
READ_LEN=100
INNER_DIST=400Consensus caller
bcftools call -c -o aln_consensus.bcf -O b aln.bcfUsing GATK for calling variants
We'll use another variant caller to call variants and compare them to the variants called by BCFtools. The HaplotypeCaller is capable of calling SNPs and indels simultaneously via local de-novo assembly of haplotypes in an active region. Firstly, download and extract GATK; you'll need to register an account and to agree to the terms and conditions.
tar -xjf GenomeAnalysisTK-3.5.tar.bz2 Then we need to setup Picard to prepare our reference fasta file:
git clone https://github.com/broadinstitute/picard.git
cd picard
git clone https://github.com/samtools/htsjdk.git
cd htsjdk
# install ant on Debian/Ubuntu
# sudo apt-get install ant
ant htsjdk-jar
cd ..
ant -lib lib/ant package-commands
cd ..Some necessary steps before running HaplotypeCaller:
java -jar picard/dist/picard.jar CreateSequenceDictionary R=test_31.fa O=test_31.dict
samtools faidx test_31.fa
# add read groups to the BAM file
java -jar picard/dist/picard.jar AddOrReplaceReadGroups \
INPUT=aln.bam \
OUTPUT=aln_rg.bam \
RGLB=test \
RGPL=illumina \
RGPU=test \
RGSM=test
# check out the header
# to see the read groups we added
./samtools view -H aln_rg.bam
@HD VN:1.5 SO:coordinate
@SQ SN:1000000 LN:1000000
@RG ID:1 LB:test PL:illumina SM:test PU:test
@PG ID:bwa PN:bwa VN:0.7.13-r1126 CL:bwa/bwa mem test_31.fa l100_n1000000_d400_31_1.fq l100_n1000000_d400_31_2.fq
# index
./samtools index aln_rg.bamNow to call variants:
java -Xmx4G -jar GenomeAnalysisTK.jar -R test_31.fa -T HaplotypeCaller -I aln_rg.bam -o aln_rg.vcfComparing VCF files
How many variants were called using BCFtools?
# convert BCF to VCF
bcftools convert -O v -o aln_consensus.vcf aln_consensus.bcf
# count
cat aln_consensus.vcf | grep -v "^#" | wc -l
9704How many variants using HaplotypeCaller?
cat aln_rg.vcf | grep -v "^#" | wc -l
9875My mutate_fasta.pl script outputs a log of the insertions, deletions, and substitutions made to a reference sequence. The pipeline stores this in the file test_mutated.log. In total there were 10,000 variants, since the mutation percent was set to 1% for a reference sequence of 1,000,000 bp.
tail test_mutated.log
999272 point: G -> C
999502 point: G -> A
999579 point: G -> T
999704 insert: G
999907 point: T -> A
999981 delete: C
Point: 3319
Delete: 3341
Insert: 3340
Total: 10000HaplotypeCaller was able to call 98.8% of the variants.
Decompose and normalise
Despite the VCF being a standard, there are still differences between VCF files. To ensure the VCF files are unified, we'll use the vt program to decompose and normalise the variants. For more information, refer to this blog post.
# download and compile
git clone https://github.com/atks/vt.git
cd vt
make
make test
cd ..
# decompose and normalise
vt/vt decompose -s aln_consensus.vcf | vt normalize -r test_31.fa - > aln_consensus.vt.vcf
# this step doesn't do anything because
# the GATK variant file is already decomposed and normalised
vt/vt decompose -s aln_rg.vcf | vt normalize -r test_31.fa - > aln_rg.vt.vcfSnpSift
We can use SnpSift to compare VCF files; I have a blog post with more information.
# download
wget http://downloads.sourceforge.net/project/snpeff/snpEff_latest_core.zip
unzip snpEff_latest_core.zipSnpSift will only compare samples with the same name, so we need to rename the sample name in one of the files to match the other. The GATK sample name was based on the read group information we added with Picard, which was test. We can use sed to change the sample name to test in the VCF file created using BCFtools.
# rename sample name to test
cat aln_consensus.vt.vcf | sed 's/\taln.bam/\ttest/' > aln_consensus.vt.renamed.vcf
# run SnpSift
java -Xmx1g -jar \
snpEff/SnpSift.jar concordance \
-v aln_consensus.vt.renamed vcf aln_rg.vt.vcf \
> concordance_by_variant.txtSnpSift will create three summary files; the concordance_aln_consensus_aln_rg.by_sample.txt file will give a sample level summary of the concordance between the two VCF files. The file is more easily viewed with the columns transposed to rows.
cat concordance_aln_consensus_aln_rg.by_sample.txt | script/transpose.pl | column -t
sample test
MISSING_ENTRY_aln_consensus/MISSING_ENTRY_aln_rg 0
MISSING_ENTRY_aln_consensus/MISSING_GT_aln_rg 0
MISSING_ENTRY_aln_consensus/REF 0
MISSING_ENTRY_aln_consensus/ALT_1 0
MISSING_ENTRY_aln_consensus/ALT_2 302
MISSING_GT_aln_consensus/MISSING_ENTRY_aln_rg 2
MISSING_GT_aln_consensus/MISSING_GT_aln_rg 0
MISSING_GT_aln_consensus/REF 0
MISSING_GT_aln_consensus/ALT_1 0
MISSING_GT_aln_consensus/ALT_2 0
REF/MISSING_ENTRY_aln_rg 0
REF/MISSING_GT_aln_rg 0
REF/REF 0
REF/ALT_1 0
REF/ALT_2 0
ALT_1/MISSING_ENTRY_aln_rg 13
ALT_1/MISSING_GT_aln_rg 0
ALT_1/REF 0
ALT_1/ALT_1 0
ALT_1/ALT_2 7
ALT_2/MISSING_ENTRY_aln_rg 118
ALT_2/MISSING_GT_aln_rg 0
ALT_2/REF 0
ALT_2/ALT_1 0
ALT_2/ALT_2 9518
ERROR 48There are five categories: MISSING_ENTRY, MISSING_GT, REF, ALT_1, and ALT_2. For each variant in each file, the genotypes are compared. (ERROR refers to incompatible variants; the REF and ALT are different) If a variant was homozygous ALT in both files, ALT_2/ALT_2 will be incremented by 1. In the table above, we see that 9,518 variants were called homozygous ALT by both variant callers. 118 variants were called homozygous ALT by BCFtools but were missing, i.e. not called by GATK. 7 variants were (mistakenly) called heterozygous by BCFtools and homozygous ALT by GATK. 13 variants were (mistakenly) called heterozygous by BCFtools and missing in the GATK VCF file. 302 variants were not called by BCFtools but were called homozygous ALT by GATK.
The concordance_by_variant.txt file will give a variant level summary. To get the column numbers of this file we can use a combination of command line tools.
cat concordance_by_variant.txt | head -1 | script/transpose.pl | nl
1 chr
2 pos
3 ref
4 alt
5 MISSING_ENTRY_aln_consensus/MISSING_ENTRY_aln_rg
6 MISSING_ENTRY_aln_consensus/MISSING_GT_aln_rg
7 MISSING_ENTRY_aln_consensus/REF
8 MISSING_ENTRY_aln_consensus/ALT_1
9 MISSING_ENTRY_aln_consensus/ALT_2
10 MISSING_GT_aln_consensus/MISSING_ENTRY_aln_rg
11 MISSING_GT_aln_consensus/MISSING_GT_aln_rg
12 MISSING_GT_aln_consensus/REF
13 MISSING_GT_aln_consensus/ALT_1
14 MISSING_GT_aln_consensus/ALT_2
15 REF/MISSING_ENTRY_aln_rg
16 REF/MISSING_GT_aln_rg
17 REF/REF
18 REF/ALT_1
19 REF/ALT_2
20 ALT_1/MISSING_ENTRY_aln_rg
21 ALT_1/MISSING_GT_aln_rg
22 ALT_1/REF
23 ALT_1/ALT_1
24 ALT_1/ALT_2
25 ALT_2/MISSING_ENTRY_aln_rg
26 ALT_2/MISSING_GT_aln_rg
27 ALT_2/REF
28 ALT_2/ALT_1
29 ALT_2/ALT_2
30 ERRORFirst 10 variants missing by GATK but called by BCFtools (column 25).
cat concordance_by_variant.txt | awk '$25 == 1 {print}' | column -t | head
1000000 13250 A AT 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 36565 T TTA 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 37667 A C 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 37668 T A 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 37670 C T 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 46727 A C 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 46729 C T 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 46730 T TC 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 48289 C G 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
1000000 48291 A G 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0First 10 variants missing by BCFtools but called by GATK (column 9).
cat concordance_by_variant.txt | awk '$9 == 1 {print}' | column -t | head
1000000 1356 TG T 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 1379 A G 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 10452 T G 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 35454 G GT 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 36563 A AT 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 37669 TC T 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 48288 A AG 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 48876 C CA 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 52138 AT A 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1000000 52141 CTA C 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0Check my mutation log.
cat test_mutated.log | awk '$1>37500 {print}' | head
37535 insert: A
37585 delete: T
37670 insert: C
37675 delete: C
37850 point: A -> C
37851 point: G -> T
37940 point: A -> G
37996 insert: T
38094 insert: A
38474 insert: TBoth tools have difficulty calling the variants (INDELs) that occur in close proximity to each other. The positions in the mutation log are slightly off because insertions and deletions were added sequentially and the positions of variants will be affected by INDEL variants added afterwards.
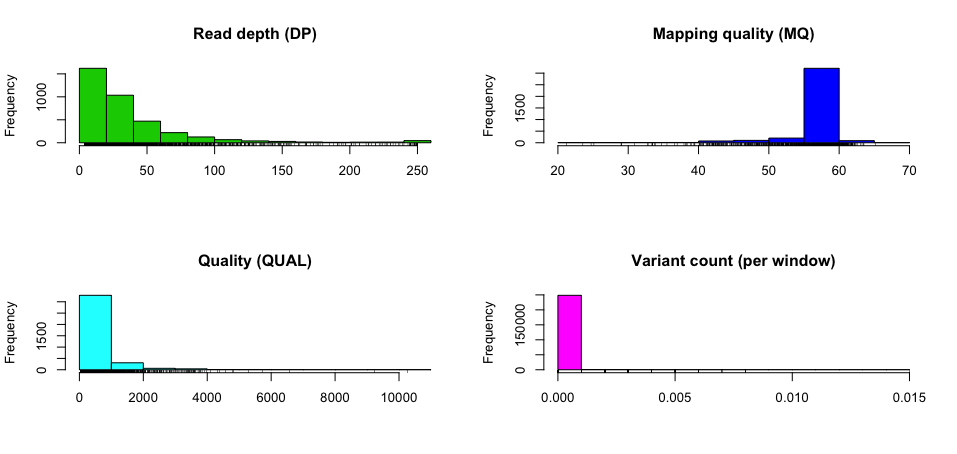
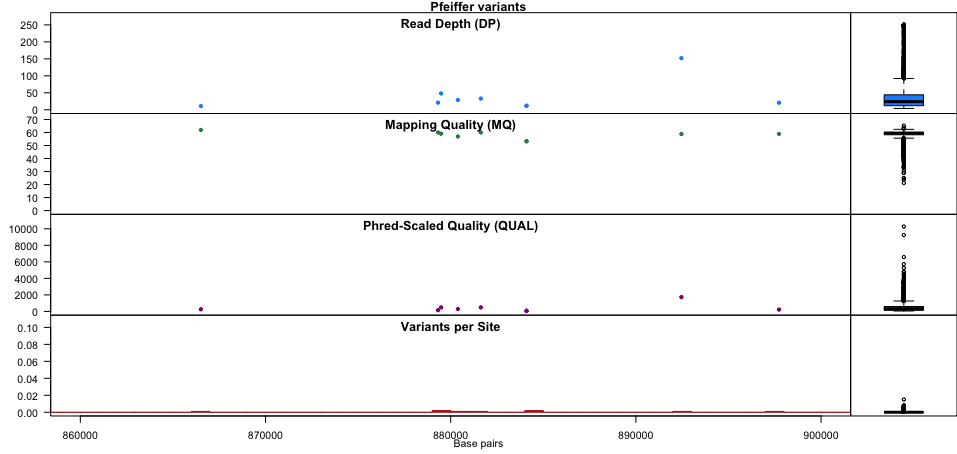
Visualisation
The vcfR package produces some nice plots.
install.packages("vcfR")
library(vcfR)
my_vcf <- read.vcfR("~/github/learning_vcf_file/eg/Pfeiffer.vcf", verbose = FALSE)
chrom <- create.chromR(name="Pfeiffer variants", vcf=my_vcf)
chrom <- proc.chromR(chrom, verbose=TRUE)
plot(chrom)chromoqc(chrom, xlim=c(860000, 900000))Useful links
- A very useful thread on SEQanswers on learning about the VCF format: http://seqanswers.com/forums/showthread.php?t=9345
- Useful tutorial on VCFs files from the 1000 Genomes Project Page: http://www.1000genomes.org/node/101
- The author of ANNOVAR writes about VCF files: http://annovar.openbioinformatics.org/en/latest/articles/VCF/
- Encoding Structural Variants in VCF version 4.0