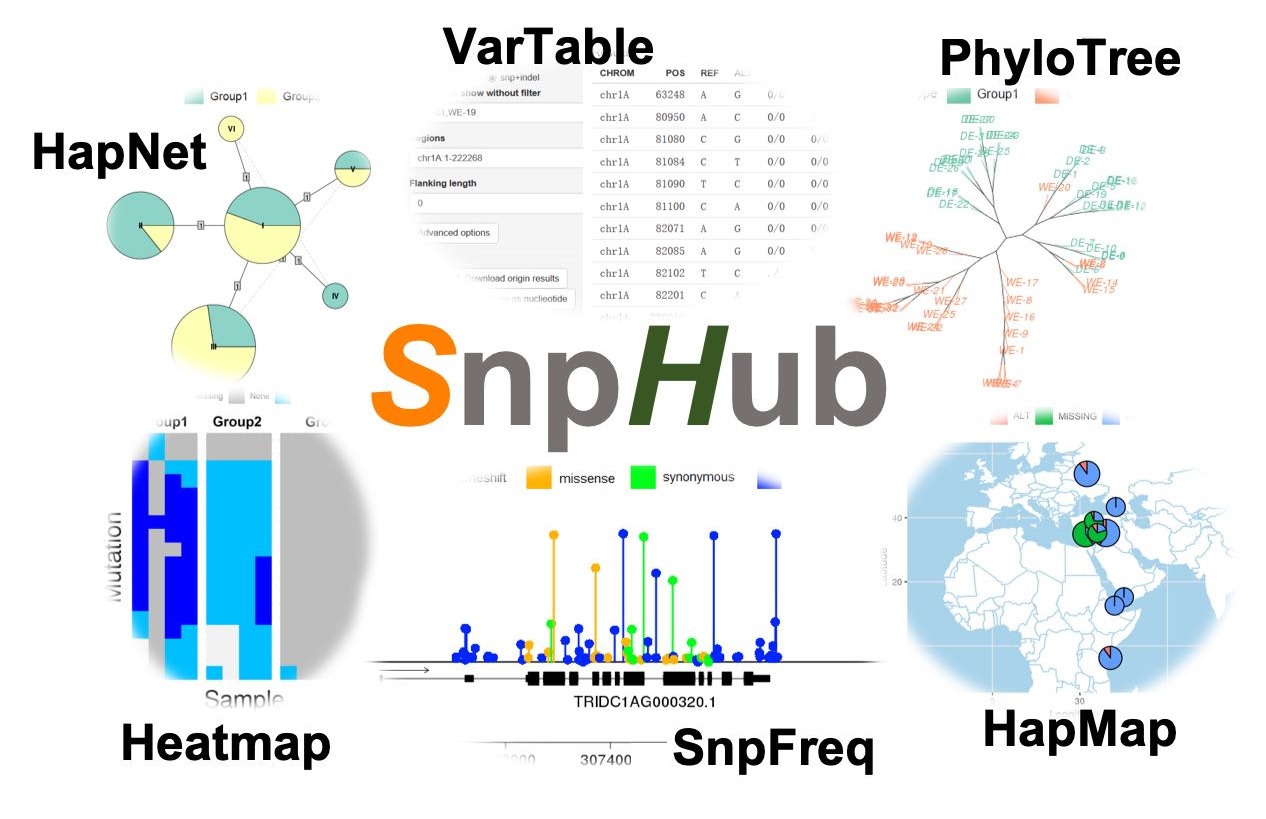
SnpHub is a Shiny-based server framework for retrieving, analyzing and visualizing the large genomic variations data in a lab.
For more details, check our tutorial
For Docker-encapsulated version, check here
To start a general setup, see here
Here are our live demos.
Please cite SnpHub as follow:
Wenxi Wang, Zihao Wang, Xintong Li, Zhongfu Ni, Zhaorong Hu, Mingming Xin, Huiru Peng, Yingyin Yao, Qixin Sun, Weilong Guo, SnpHub: an easy-to-set-up web server framework for exploring large-scale genomic variation data in the post-genomic era with applications in wheat, GigaScience, Volume 9, Issue 6, June 2020, giaa060, https://doi.org/10.1093/gigascience/giaa060
To run the SnpHub, make sure the following software programs are already installed:
- samtools (≥ v1.4)
- bcftools (≥ v1.8)
- seqkit
- tabix (≥ v1.6)
Also, the following R packages are also prerequisites:
- ggplot2
- crayon
- ggmap
- dplyr
- rjson
- shiny
- pegas
- maps
- vcfR
- ape
- DT
Two steps are needed to run on demo data set:
- Using
gitto clone SnpHub to local.
git clone https://github.com/esctrionsit/snphub- Using
Rto run on demo data set. (Or copy the SnpHub into yourshiny-server app folder)
R -e "shiny::runApp('./snphub', port=5000, host='0.0.0.0')"Make sure that samtools, bcftools, seqkit and tabix are added to system PATH. Otherwise, tool application paths part in setup.conf is needed to change to fit.
Basic config file is named setup.conf, fulfill it and then run command:
Rscript ./setup.RCheck configuration for more details about setup.conf.
Check file format for more details about required file formats.
If it is your first time to set up SnpHub, you need to delete advanced_config.R, and then rename advanced_config_O.R to advanced_config.R.
For more details, check our tutorial