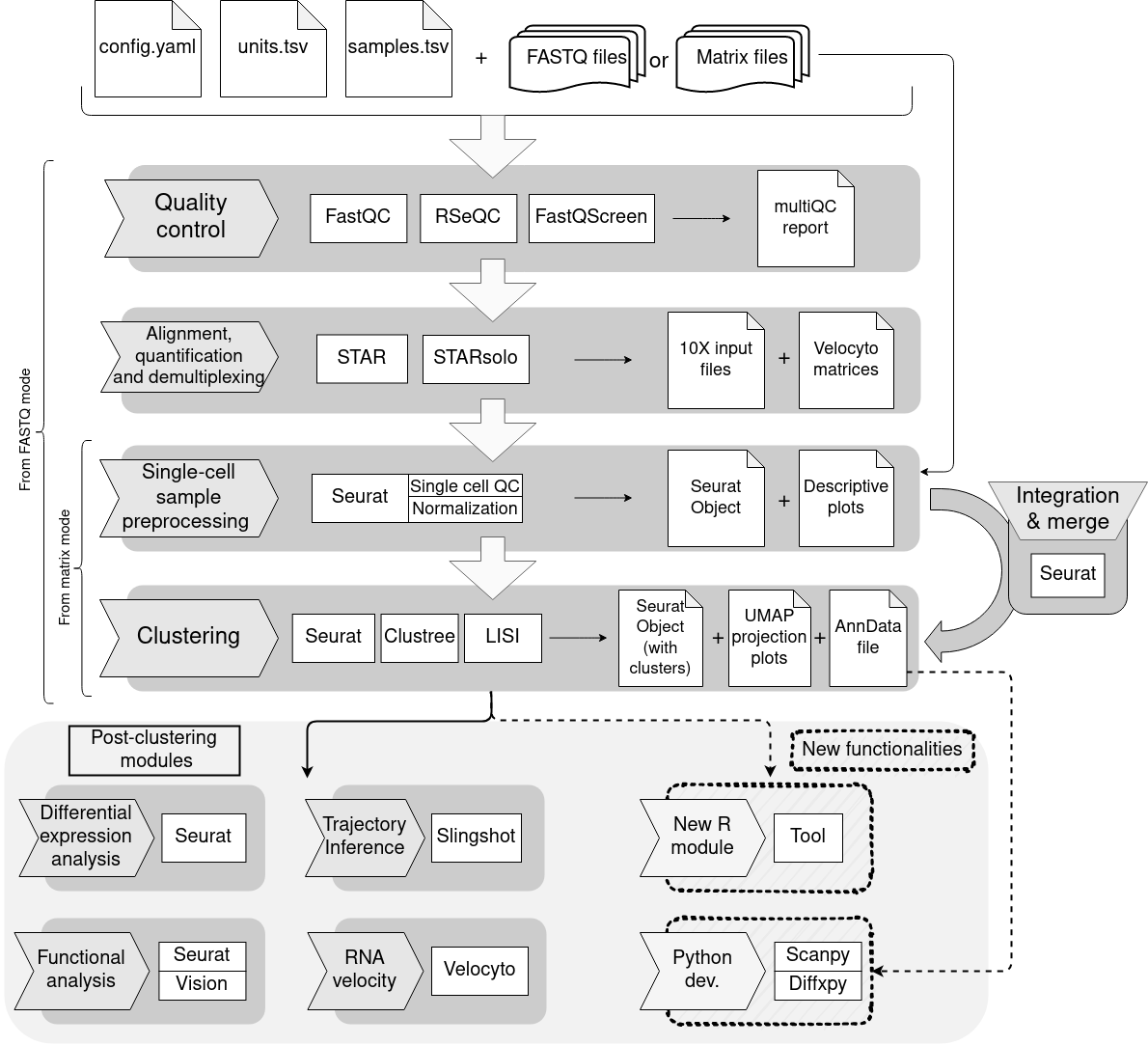
bollito is a Snakemake pipeline that performs a comprehensive single-cell RNA-seq analysis, covering both the basic steps (QC, alignment, quantification and cell specific QC) and more advanced downstream analyses (clustering, diferential expresion, trajectory inference, functional analysis and RNA velocity).
This pipeline uses state-of-the-art single-cell RNA-seq tools like Seurat, STARsolo, Vision, Slingshot, and Velocyto.
The pipeline makes extensive use of Snakemake's integration with the conda package manager and docker/singularity containers, to automatically take care of software requirements and dependencies.
bollito has two main modes of execution depending on the input data:
- From FASTQ: it accepts FASTQ-formatted raw data (from drop-seq or 10x Genomics experiments).
- From matrices including:
- Feature-barcode matrices (expression matrix format from STARsolo).
- Standard matrices (cell names and gene names included in the matrix).
We've built bollito for flexibility, with the aim of allowing the user to adjust the pipeline to different experiments using configuration parameters. This includes adjusting the cell filtering, normalization, variables regression, number of significant components, clustering resolution, etc. When in doubt, the default parameters were calculated to offer a good starting configuration. Additionally, once the main steps of the pipeline are finished, bollito creates a AnnData file from the Seurat file and test if it is correctly formatted. This feature improves the interconnection between R and Python, helping users which might want to perform analysis with tools from both languages.
This is a schema of the complete workflow. Certain parts may be skipped depending on input data and chosen configuration.
- Luis García-Jimeno
- Coral Fustero-Torre
- María José Jiménez-Santos
- Gonzalo Gómez-López
- Tomás Di Domenico
- Fátima Al-Shahrour
The setup of the pipeline consists in the modification of three configuration files, indicating the desired parameters and the location of the input files. A general description of these files follows. See the Usage section for more details.
- config.yaml contains all pipeline parameters.
- samples.tsv contains information on the samples to be analysed.
- units.tsv: contains information on the different data files associated to the samples to be analysed.
- raw data in gzip compressed FASTQ files
or
-
matrices of count data
- 10x-like input (matrix.mtx + genes.tsv + barcodes.tsv)
or
- standard tabular format (where rows are genes and columns are cells)
bollito requires the conda package manager in order to work. If you have singularity installed and would rather not install conda, you can provide the "--use-singularity" option when running the pipeline (see below). Otherwise, please install conda by following the bioconda installation instructions.
Use git clone command to create a local copy.
git clone https://gitlab.com/bu_cnio/bollito.git
Before executing the pipeline, the users must configure it according to their samples. To do this, they must fill these files:
TIP: different analysis can be run using just one cloned repository. This is achieved by changing the outdir and logdir in the configuration file. Also different parameters values can be used in the different analysis.
This file contains information on the samples to be analyzed. The first column, called "sample", is mandatory, and defines the sample name for each sample. The other columns are used to define the samples. This information is stored as metadata in the Seurat object, so, every cell belonging to that sample is labeled with the value defined by the user, meanwhile the column heaeder will correspond to the condition name.
An example file (samples-example.tsv) is included in the repository.
Rename it to samples.tsv and edit its contents to list the sample names and the features
related to each sample.
This file is used to configure the input files.
There are two example files, depending on the type of input data:
- If your input are FASTQ files:
An example file (units-example_fastq.tsv) is included in the repository.
Rename it to units.tsv and edit its contents according to the following table:
| Field name | Description |
|---|---|
| sample | Sample name (must match the sample name specified in samples.tsv). |
| unit | A distinct name for each pair of files associated to the same sample (for example in the case of replicates). |
| fq1 | FASTQ file for read 1, containing the Cell Barcode and UMI. |
| fq2 | FASTQ file for read 2, containing the transcriptomic sequence. |
- If your input are matrix files:
An example file (units-example_matrices.tsv) is included in the repository.
Rename it to units.tsv and edit its contents according to the following table:
| Field name | Description |
|---|---|
| sample | Sample name (must match the sample name specified in samples.tsv). |
| matrix | Matrix file (.mtx for 10x or .tsv for standard) storing the counts. |
| cell_names (10x only) | tsv file containing one cell name per row. |
| gene_names (10x only) | tsv file containing one gene name per row. |
| metadata (optional) | tsv file with two or more columns. First column corresponds to each cell name specified in cell_names.tsv and the rest are metadata variables. First row indicates the metadata variable name. |
This is the pipeline configuration file, where you can tune all the available parameters to customise your single-cell analysis.
The example file (config-example.yaml) features extensive inline documentation.
Here are some of the main available parameters:
| Parameter | Description |
|---|---|
| input_type | Type of input data (fastq or matrix). |
| technology | Technology used to get the reads files (10x or Drop-seq). |
| outdir | Directory where to store the output files. |
| logdir | Directory where to store the log files. |
| graphics | Graphic card availability. |
| random_seed | Seed parameter to allow for reproducible analyses. |
| case | Type of case used to represent the gene names, must be use according the specie and genesets used. |
| annotation | GTF file holding genetic features information. |
| idx | Folder containing STAR genomes indexes. |
| whitelist | Cell barcodes whitelist file needed for 10x experiments quantification and demultiplexing. |
This file is optional and it is only used when the input file used are matrices. The purpose of this file is to annotate each individual cell, storing that information in the Seurat object. It is a tsv file with two or more columns. The first column corresponds to each cell name specified in cell_names.tsv and the rest are the values of the metadata variables. First row of each column indicates the metadata variable name.
To run the pipeline, the user needs to create the conda environments first, which will take some minutes. This step is done automatically using this command:
snakemake --use-conda --conda-create-envs-only --conda-frontend mamba
Once the pipeline is configured and conda environments are created, the user just needs to run bollito.
snakemake --use-conda -j 32
The mandatory arguments are:
- --use-conda: to install and use the conda environemnts.
- -j: number of threads/jobs provided to snakemake.
Here you can find a general description of the main steps of the pipeline.
bollito implements FastQC to check the overal quality of the input FASTQ files.
FastQ Screen can optionally be enabled in order to check the input FASTQ files for contaminants.
bollito creates a Quality Control HTML report using MultiQC. This report includes information from FastQC and RSeQC (which will be explained later).
To obtain the cell expression profiles we need to align the FASTQ files against an annotated reference genome. Once we obtain the aligned files, the pipeline assigns the reads to their corresponding cell barcodes, and performs an UMI-based quantification of the annotated features.
The alignment tool implemented in bollito is STAR, taking advantage of its STARsolo mode for single-cell RNA-seq quantification.
This step requires the following parameters, defined in the configuration file:
- An annotation file containing the features of interest (in GTF format, must match the target genome)
- One of the following options for the genome:
- a FASTA file (will be indexed by bollito).
- a STAR genome index.
- Technology, which includes Drop-seq, 10x (chemistry version also needs to be specified), or a custom technology where the user will need to set the CB and UMI length.
- The corresponding CB whitelist.
Example parameters for different STAR configurations are available in the example config file.
Quality control of the resulting alignments is performed using RSeQC.
Once the UMI-count matrix is obtained, bollito performs a cell-based quality control. The purpose of this step is to control for broken cells and doublets, and to analyze the cell cycle status of the cells. Next, the normalization and dimensionality reduction steps are applied.
Cell quality control, normalization, and dimensionality reduction are implemented using the Seurat package.
Cell cycle checking is implemented based on molecular signatures from Tirosh et al, 2015.
In order to customize this step, the following parameters can be adjusted via the configuration file:
- Minimum and maximum number of detected genes per cell.
- Minimum and maximum read counts per cell.
- Minimum ribosomal content.
- Minimum mitochondrial content.
- Normalization method "SCT" ((Hafemeister & Satija, 2019)) or "standard".
The normalization itself can also be parametrized, including the possibility of regressing the desired variables (such as cell cycle scoring, number of detected genes, % of mitochondrial genes) in order to mitigate their effect on the dataset. Refer to the example config file for the available options.
NOTE: the user will have to choose the correct value for some parameters (QC threshold filters, significant PCA components or regressed variables) after the execution of these steps. For this purpose, it is necessary to stop the pipeline executions when these steps are finished. Then, take a look and the results to define those parameter values at the configuration file. Finally, re-run the pipeline from these steps in order to apply the changes.
bollito can perform an optional merging step. This step consists in combining all the sample's Seurat objects which come from the single-cell QC step. No normalization is performed in this step, since the merged object will be used as input in the normalization step.
To enable this step, the user just need to set the "enabled" parameter to TRUE in the configuration file.
bollito allows for an optional integration step, in order to detect shared cell states between datasets. The integration method is based on the identification of anchor cells between the datasets, and the projection of datasets on each other by using these anchors. For more information regarding the integration step, please refer to the Seurat Integration and Label Transfer documentation.
Integration step also performs the normalization of the integrated samples. It uses the same normalization method and values specified for the normalization rule, so the user does not need to specify any extra parameter.
To enable this step, the user just need to set the "enabled" parameter to TRUE in the configuration file.
Clustering of cells uses the normalized expression profiles. After, a dimensionality reduction by PCA, the k-nearest-neightbor (KNN) graph embedded in this lower dimensional space, is obtained. Then a Shared Nearest Neighbour (SNN) graph is constructed calculating the nearest neighbour cells overlapping using the Jaccard index. Once the graph is created, clusters are captured by using a the Louvain algorithm.
To explore the clusters along the resolutions, bollito uses Clustree. Cluster validation is achieved by calculating silhouette scores for each cluster. LISI ((Korsunsky, I. et al.) is used to assess if there is a batch effect produced by any of the categorical varaibles that are described in the experiment. If any batch effect is detected I would be advisabe to regress out that variable.
Additionally, once the main steps of the pipeline are finished, bollito creates a AnnData file from the Seurat output file and checks its formatting.
For this step, the following parameters need to be adjusted via the configuration file:
- Number of significant components based on the elbow plot or JackStraw analysis obtained in previous steps.
- Number of neighbours (k) used to generate the KNN graph (default = 20).
- Resolutions to be used in the community detection method.
NOTE: the best resolution obtained should be specified in the configuration file, since it is used in posterior effects.
Differential expression analysis is based on the condition that the user requires, including a specific cluster resolution or some annotation information. For each condition, two analyses are performed:
- Marker gene detection (for this test, only genes with a logFC threshold of 0.25, that are expressed in at least 10% of the cells are included).
- Differential expression for all genes (no thresholds applied).
Output from this step includes a heatmap of top marker genes per condition, and a .rnk file that may be used for a downstream gene enrichment analysis with GSEA,
The following parameters of this step need to be adjusted via the configuration file:
- Set enabled to True.
- Condition to analyze (cluster resolution or annotation information).
- Differential expression test to apply.
For a list of available tests see the Seurat FindMarkers function documentation.
bollito uses Seurat to apply the AddModuleScore function to the molecular signatures specified by the user. A paired Wilcoxon test is applied to compare the expression of the genes from the signature between the clusters.
To enable this step, the following parameters need to be adjusted via the configuration file:
- Set enabled to True.
- Set the path to the molecular signatures file to use (in .gmt format).
- Set the ratio of expressed genes / total genes from a geneset to be tested (default is 0.2).
bollito applies the Vision methodology in order to study different molecular signatures and their significance at a specific clustering resolution.
To enable this step, the following parameters need to be adjusted via the configuration file:
- Set enabled to True.
- Set the path to the molecular signatures file to use (in .gmt format).
- Select the desired metadata variables from Seurat.
- Set the desired cluster resolution.
This step analyses the cell lineages of your sample by inferring a pseudotime variable from the data and sorting the clusters according to it. The trajectory inference step is implemented by using the Slingshot package.
To enable it, the following parameters need to be adjusted via the configuration file:
- Set enabled to True.
- Cluster resolution must be specified.
- Specifiy start and end cluster(s) (optional).
- Number of genes for the heatmap representation.
- Number of most variable genes (allows you to generate the general additive model of the heatmap).
The analysis of RNA velocity allows you to capture the expression dynamics of your data by estimating the spliced and unspliced mRNA abundances on each of the available splicing sites. Based on this information, future state of single-cells can be inferred. This step is implemented using the Velocyto wrapper.
To enable this step, the following parameters need to be adjusted via the configuration file:
- Set enabled to True.
- Cluster resolution must be specified.
- If the dataset is large, a downsampling should be considered (optional).
NOTE: RNA velocity step can not be performed if we use a matrix as input of the pipeline, since it needs the BAM files to generate the three count matrices (spliced, unspliced and ambiguous).
The user can configure bollito to optimise the available computational resources in order to reduce the computational time. The optimization is achieved thanks to Snakemake's ability to run several samples at the same time and single-cell script parallelisation using the future package implementation. Resources configuration is done through the configuration file. This file has a field called resources, where the user can define the RAM memory usage and the number of threads (if the rule admits multithreading) available to a specific step. Additionally, if the user does not provide any value for some of these fields, the pipeline will use the default values.
bollito features a shortcut system based on specfic targets related to the pipeline's steps. Each target calls a end point rule which terminate the pipeline execution.
To use the shorcuts, you only need to run the pipeline as usual, but with the --until option.
snakemake --use-conda --until target_name
The available targets are:
- expression_matrix: run bollito until alignment step included.
- qc_expression_matrix: run bollito until single-cell QC step included (rules: seurat_qc, seurat_postqc & seurat_merge).
- normalized_expression_matrix: run bollito until single-cell normalization step included (rules: seurat_qc, seurat_postqc, seurat_merge, seurat_normalization & seurat_integration).
Additionally, the user might use the Snakemake rules names as targets, which are available in the config.yaml file.
bollito produces a HTML report using Snakemake's automatic report generaration. This report includes the multiQC report and some quality control and normalization information at single-cell level from Seurat.
To generate the report, you only need to use --report option when the analysis is finished.
snakemake --report report.zip
This will generate a zip file containing an HTML index page and all the files needed to correctly display the report.
Bollito generates an AnnData output file to allow users to perform downstream analyses using Scanpy and other python-based packages. This AnnData file is obtained from the post-clustering Seurat object, so it stores all the annotations and cell filterings applied until that step.
The following metrics were generating using the 10K PBMC 3p from 10x Next GEM Chromium X dataset on a HPC cluster running CentOS 8 (248 cores, 2Tb RAM).
| pipeline step | running time (s) | max RAM usage (MB) | threads |
|---|---|---|---|
| fastqc | 657.897 | 3830.980 | 4 |
| star | 5388.925 | 36103.887 | 8 |
| rseqc_junction_saturation | 1310.431 | 4848.223 | 1 |
| rseqc_readdis | 2507.294 | 1058.127 | 1 |
| rseqc_stat | 1022.082 | 116.960 | 1 |
| rseqc_readgc | 1166.199 | 2183.963 | 1 |
| rseqc_readdup | 2123.237 | 31723.523 | 1 |
| rseqc_infer | 7.545 | 150.740 | 1 |
| rseqc_innerdis | 630.251 | 970.620 | 1 |
| rseqc_junction_annotation | 1171.999 | 291.133 | 1 |
| multiQC | 40.741 | 212.860 | 2 |
| seurat_preQC | 120.221 | 2310.437 | 1 |
| seurat_postQC | 65.062 | 1869.23 | 1 |
| seurat_normalization | 405.939 | 9125.253 | 1 |
| seurat_find-clusters | 253.288 | 3710.987 | 2 |
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc [Accessed 13 March 2020]
- Wingett S. (2010). FastQ Screem:FastQ Screen allows you to screen a library of sequences in FastQ format against a set of sequence databases so you can see if the composition of the library matches with what you expect. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen [Accessed 13 March 2020]
- Dobin, A., Davis, C., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T., 2012. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), pp.15-21.
- Wang, L., Wang, S. and Li, W., 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics, 28(16), pp.2184-2185.
- Kowalczyk, M., Tirosh, I., Heckl, D., Ebert, B. and Regev, A., 2014. Single cell RNA-Seq of hematopoietic stem cells reveals a cell cycle-dependent interplay between aging and differentiation. Experimental Hematology, 42(8), p.S21.
- Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W., Hao, Y., Stoeckius, M., Smibert, P. and Satija, R., 2019. Comprehensive Integration of Single-Cell Data. Cell, 177(7), pp.1888-1902.e21.
- Hafemeister, C. and Satija, R., 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology, 20(1).
- Zappia, L. and Oshlack, A., 2018. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience, 7(7).
- DeTomaso, D., Jones, M., Subramaniam, M., Ashuach, T., Ye, C. and Yosef, N., 2019. Functional interpretation of single cell similarity maps. Nature Communications, 10(1).
- Street, K., Risso, D., Fletcher, R., Das, D., Ngai, J., Yosef, N., Purdom, E. and Dudoit, S., 2018. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics, 19(1).
- La Manno, G., Soldatov, R., Zeisel, A., Braun, E., Hochgerner, H., Petukhov, V., Lidschreiber, K., Kastriti, M., Lönnerberg, P., Furlan, A., Fan, J., Borm, L., Liu, Z., van Bruggen, D., Guo, J., He, X., Barker, R., Sundström, E., Castelo-Branco, G., Cramer, P., Adameyko, I., Linnarsson, S. and Kharchenko, P., 2018. RNA velocity of single cells. Nature, 560(7719), pp.494-498.
- Korsunsky, I., Millard, N., Fan, J., Slowikowski, K., Zhang, F., & Wei, K., Baglaenko, Y., Brenner, M., Loh, P. and Raychaudhuri, S. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods, 16(12), pp.1289-1296.
The system is pre-configured to run an example based on sample data available from 10x Genomics. The required datasets can be found at these URLS. Please update the "units.tsv" file to point at the data as needed.
- https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/neuron_10k_v3
- https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/heart_10k_v3
The pipeline can optionally run FASTQ_SCREEN to check the samples for contamination.
To disable it use the config file option config["rules"]["fastq_screen"]["disabled"].
Config file pointing to indexes should be placed in a directory named config['rules']['fastq_screen_indexes']['outdir']/FastQ_Screen_Genomes.
If the rule is enabled and no config file provided, default indexes will be downloaded using the command fastq_screen --get_genomes.