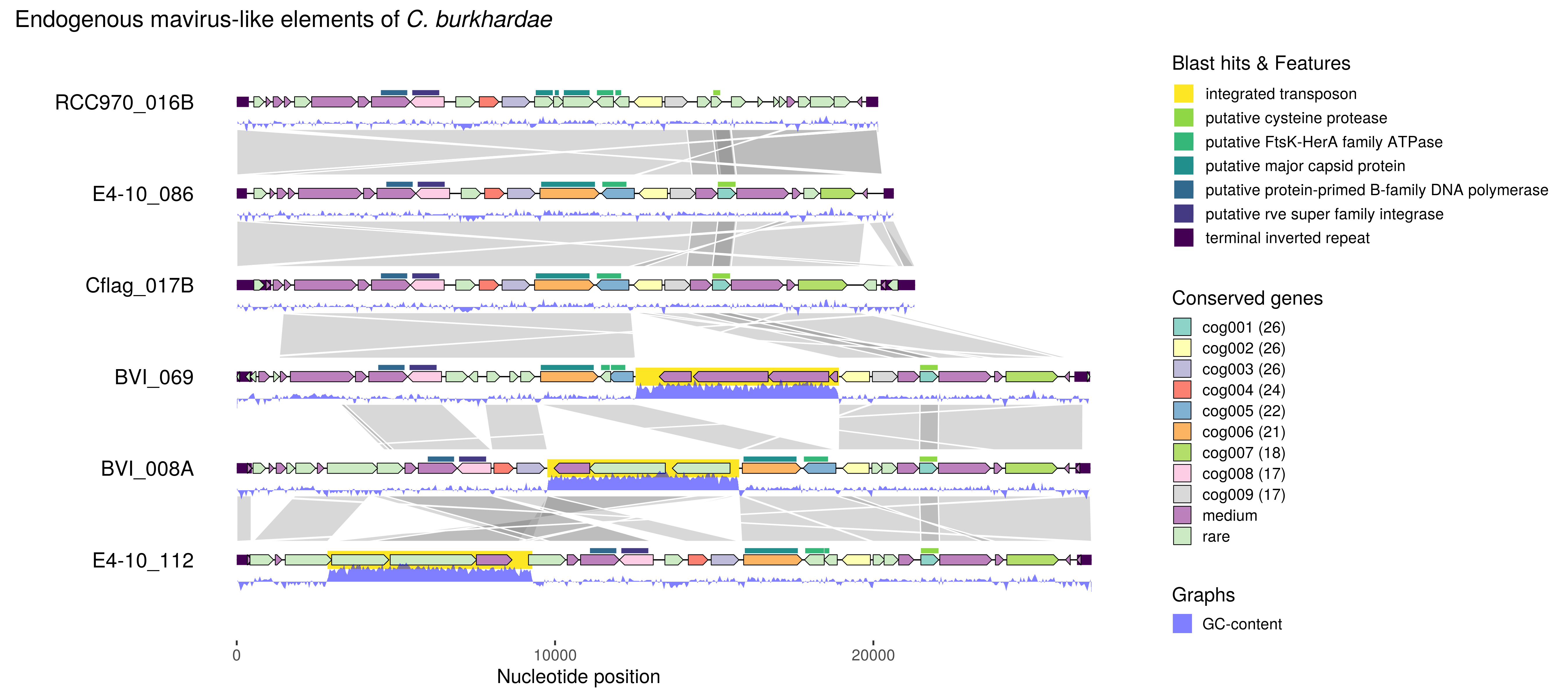
gggenomes is a versatile graphics package for creating comparative genomic maps, extending the popular R visualization package ggplot2. It adds dedicated plot functions (geoms, positions) for common sequence and alignment features, such as gene models and syntenic regions, as we well as verbs to further manipulate the plot and the underlying data, for example, to quickly zoom in into gene neighborhoods.
For a reproducible recipe describing the full evolution of this plot starting from a mere set of contigs, and including the bioinformatics analysis workflow, have a look at From a few sequences to a complex map in minutes. For a compact version to of the code see below.
# EMALEs: endogenous mavirus-like elements (Hackl et al. in prep.)
# a use case data bundled with ggggenomes
seqs <- emale_seqs[1:6,] # first 6 genomes only
p <- gggenomes(seqs, emale_genes, emale_tirs, emale_links) %>%
# standard tracks
add_features(emale_transposons, emale_gc) %>%
# more features
add_subfeatures(genes, emale_blast, transform="aa2nuc") %>%
# features that map onto features
add_clusters(genes, emale_cogs) # and some gene cluster info
# add geoms and customize the plot
p <- p +
geom_seq() + geom_bin_label() + # chromosomes and labels
geom_link(offset = c(0.3, 0.2), color="white", alpha=.3) +
# synthenic regions
geom_feature(aes(color="terminal inverted repeat"), use_features(2),
size=4) + # repeats
geom_feature(aes(color="integrated transposon"),
use_features(emale_transposons), size=7) +
# transposons
geom_gene(aes(fill=cluster_label)) + # colored genes
geom_feature(aes(color=blast_desc), use_features(emale_blast),
size=2, position="pile") + # some blast hits
geom_ribbon(aes(x=(x+xend)/2, ymax=y+.24, ymin=y+.38-(.4*score),
group=seq_id, linetype="GC-content"), use_features(emale_gc),
fill="blue", alpha=.5) + # combine with ggplot2 geoms
scale_fill_brewer("Conserved genes", palette="Set3") +
scale_color_viridis_d("Blast hits & Features", direction = -1) +
scale_linetype("Graphs") +
ggtitle(expression(paste("Endogenous mavirus-like elements of ",
italic("C. burkhardae"))))
# reverse-complement 3 genomes for better alignment
p <- p %>% flip_bins(3:5)
pVisualization is a corner stone of both exploratory analysis and science communication. Bioinformatics workflows, unfortunately, tend to generate a plethora of data products often in adventurous formats making it quite difficult to integrate and co-visualize the results. Instead of trying to cater to the all these different formats explicitly, gggenomes embraces the simple tidyverse-inspired credo:
- Any data set can be transformed into one (or a few) tidy data tables
- Any data set in a tidy data table can be easily and elegantly visualized
As a result gggenomes helps bridge the gap between data generation, visual exploration, interpretation and communication, thereby accelerating biological research.
Under the hood gggenomes uses a light-weight track system to accommodate a mix of related data sets, essentially implementing ggplot2 with multiple tidy tables instead of just one. The data in the different tables are tied together through a global genome layout that is automatically computed from the input and defines the positions of genomic sequences (chromosome/contigs) and their associated features in the plot.
gggenomes stands on the shoulder of giants. It was born out of admiration of David Wilkins' gggenes package, draws from other ggplot2 extensions such as Guangchuang Yu's ggtree, and is fundamentally inspired by Thomas Lin Pedersen's incredibly rich ggraph package.
gggenomes is at this point still in an alpha release state, and therefoe only available as developmental package.
# install ggtree
# https://bioconductor.org/packages/release/bioc/html/ggtree.html
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("ggtree")
# install.packages("devtools")
devtools::install_github("thackl/thacklr")
devtools::install_github("thackl/gggenomes")