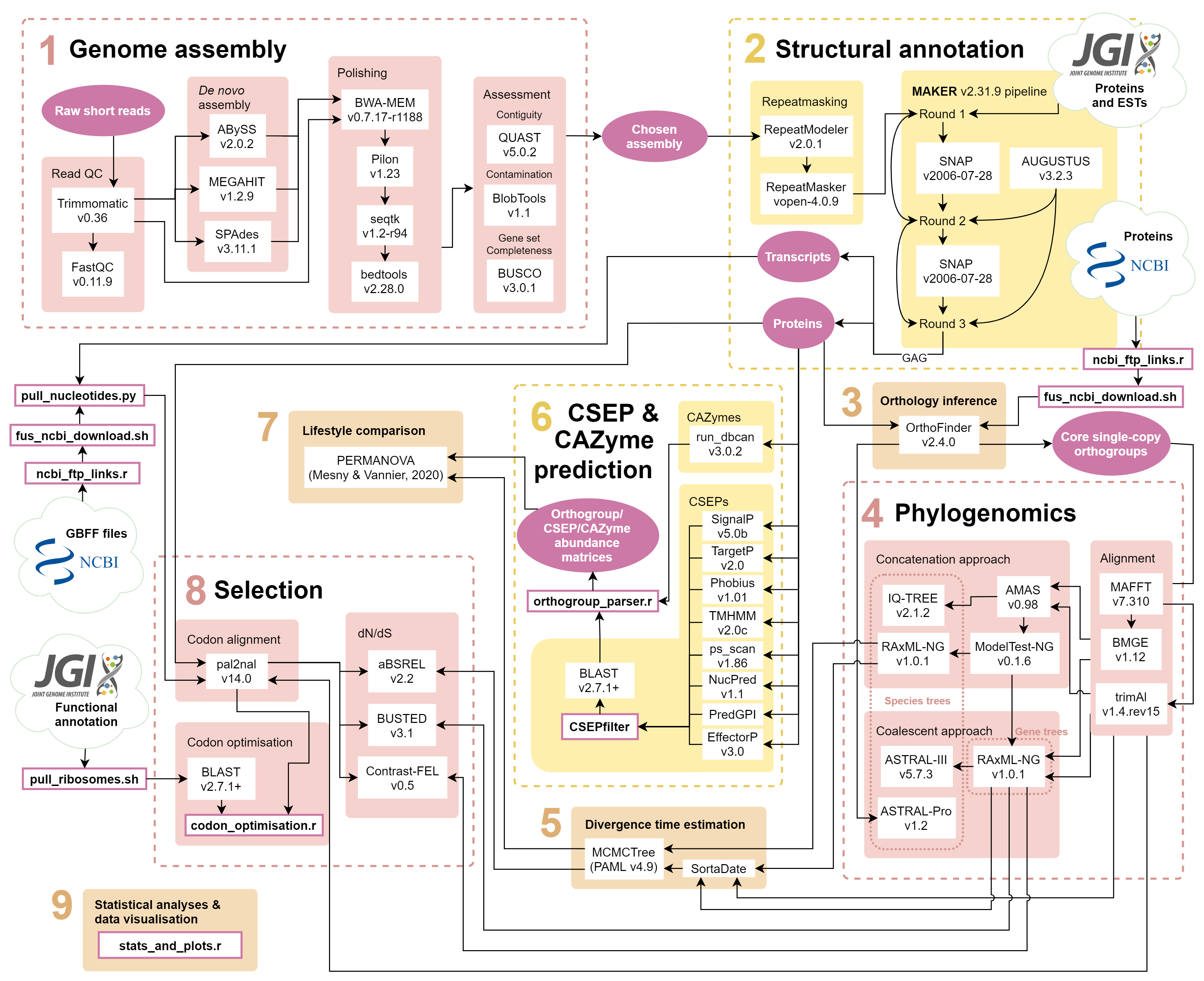
Bioinformatics analysis pipeline for:
Hill et al. (2022) Lifestyle transitions in fusarioid fungi are frequent and lack clear genomic signatures. Molecular Biology and Evolution 39(4):msac085. doi:10.1093/molbev/msac085.
The pipeline was written for and run on Queen Mary University of London's Apocrita HPC facility which uses the Univa Grid Engine batch-queue system. This means that many of the bash scripts (.sh file endings) specify core allocation, run times and memory usage allocation that may need to be adapted for different platforms.
cd assembly
cd assembly/reads
Requires raw fastq.gz paired-end reads in this directory as well as TruSeq3-PE.fa file with adapter sequences downloaded from here (for Illumina NovaSeq 6000 151bp paired-end reads).
qsub trimmomatic.sh- trims raw reads using Trimmomatic.qsub fastqc.sh- after trimming, checks read quality with FastQC.
cd assembly/denovo_assembly
./submit_assembly.sh- makes new directory and submits job scripts for each assembly tool -abyss.sh(ABySS),megahit.sh(MEGAHIT) andspades.sh(SPAdes)../abyss_comp.sh- compares the assembly stats to choose 'best' kmer size for ABySS (must be done afterabyss.shhas finished for all kmer sizes and strains).
cd assembly/polishing
1.qsub polish.sh - for each strain and assembly tool, maps raw reads to assembly and calculates mapping statistics with BWA-MEM and SAMtools and polishes the assembly with Pilon. Also removes sequences <200bp using Seqtk for NCBI compliance.
After completing 4 Assessment and uploading to NCBI:
2../ncbi_filter.sh - removes sequences identified as mitochondrial or duplicates by NCBI (listed in files saved as duplicates_* and mito_remove_* for each strain) and trims sequences identified as having mitochondrial contaminants (listed in files saved as mito_trim_*.bed) using bedtools.
cd assembly/assessment
./submit_assessment.sh- submitsquast.sh(QUAST),busco.sh(BUSCO) andblast.sh(BLAST) scripts for assembly quality statistics.busco.shrequires the Hypocreales BUSCO dataset downloaded from here).qsub blobtools.sh- submitsblobtools.shto run BlobTools (must be done afterblast.shhas finished for all strains).
cd annotation
cd annotation/repeat_masking
qsub repeatmodeler.sh- makes custom repeat library for each strain using RepeatModeler.qsub repeatmasker.sh- uses the custom repeat libraries to softmask assemblies using RepeatMasker.
cd annotation/maker
Informed by this tutorial. Requires ESTs and proteins from Fusoxy1 and Fuseq1 (Mesny et al. 2021) downloaded from Mycocosm in this directory.
cd annotation/maker/round1
qsub maker.sh - submits first run of MAKER using ESTs and proteins, as indicated in .ctl files.
cd annotation/maker/round2
qsub training_snap/snap.sh- trains SNAP using gene models from the first MAKER round.qsub maker2.sh- submits second run of MAKER using trained SNAP (as indicated in.ctlfiles).
cd annotation/maker/round3
qsub training_snap2/snap2.sh- trains SNAP again using gene models from the second MAKER round.qsub maker3.sh- submits third run of MAKER using second trained SNAP (as indicated in.ctlfiles).qsub rename.sh- after obtaining unique locus tags from e.g. NCBI (seelocus_tags.txt), renames IDs in gff and fasta files.qsub gag.sh- runs GAG to remove introns <10bp, remove terminal Ns and correct start and stop codons in gff file for NCBI compliance.
cd orthology_inference
./submit_protein_download.sh- submitsncbi_ftp_links.randprotein_download.shto download of predicted protein sets of Fusarium strains from NCBI.qsub orthofinder.sh- submits orthology inference using OrthoFinder.
cd phylogenomics
./submit_alignment.sh- submitsalignment.shfor alignment of single copy orthogroups from OrthoFinder with MAFFT followed by trimming with BMGE and trimAl../concat.sh- concatenate single copy orthogroup alignments and prepare partition files using AMAS../submit_modeltestng.sh- submitsmodeltest-ng/modeltestng.shto run ModelTest-NG on all single copy orthogroups (in computationally tractable chunks)../submit_speciestrees_concatenation.sh- submits concatenation-based species tree methods -species_tree/raxmlng.sh(RAxML-NG) andspecies_tree/iqtree.sh(IQ-TREE)../submit_RAxML-NG_genetrees.sh- submitsRAxMLNG_genetrees.shto run RAxML-NG for individual gene trees../submit_speciestrees_coalescent.sh- submits coalescent-based species tree methods -species_tree/astral.sh(ASTRAL-III) andspecies_tree/astral-pro.sh(ASTRAL-Pro) using genes trees.
cd divergence_time_estimation
cd divergence_time_estimation/sortadata
qsub sortadate - reroots gene and RAxML-NG species tree and runs with SortaDate to filter for top ten 'clock-like' genes.
cd divergence_time_estimation/mcmctree
qsub mcmctree_dating_step1.sh- adds secondary time calibrations to species tree nodes and submits first step of approximate likelihood divergence time estimation with protein data using MCMCTree from PAML (see tutorial).Rscript estimate_rate.r- estimates the scaling parameter for the substitution rate prior to be added tomcmctree_step2_independent.ctlandmcmctree_step2_correlated.ctl../submit_mcmctree_dating_step2.sh- submitsmcmctree_independent.shandmcmctree_correlated.shfor second step of approximate likelihood estimation for both independent and correlated rates relaxed clock models.
cd CSEP_CAZyme_prediction
./submit_CSEPprediction.sh- submits all programmes in the CSEP prediction pipeline -signalp/signalp.sh(SignalP),targetp/targetp.sh(TargetP),phobius/phobius.sh(Phobius),tmhmm/tmhmm.sh(TMHMM),prosite/ps_scan.sh(ps_scan),nucpred/nucpred.sh(NucPred),predgpi/predgpi.shwhich in turn submitspredgpi/PredGPI.rto use the R package ragp (PredGPI) andeffectorp/effectorp.sh(EffectorP)../submit_CSEPfilter.sh- submitsCSEPfilterto produce lists of CSEPs from all programme results../submit_CSEPblast.sh- submitsblastp/blastp.shto BLAST of CSEPs against the PHI-base database (requiresphi-base_current.csvandphi-base_current.fasto be downloaded from here into theblastpdirectory)../submit_CAZymeprediction.sh- submitsrun_dbcan/run_dbcan.shto run run_dbcan.qsub submit_orthogroupparsing.sh- submitsorthogroup_parser.rto make abundance matrices of orthogroups for all strains and categorises whether they are CSEPs/CAZymes and core/accessory/specific.
cd lifestyle_comparison
./submit_lifestyletest.sh - submits lifestyle_v_phylogeny.r to prepare input file for lifestyle-test.sh which runs PERMANOVA-based lifestyle test on orthogroup and CSEP presence absence matrices; run_edited.py is modified from the original script run.py by Mesny & Vannier.
cd selection
qsub gbff_files/ncbi_gbff_download.sh- downloads GBFF files for the strains used in this study from NCBI; also need Ilysp1 transcripts downloaded from Mycocosm ingbff_filesdirectory../submit_pal2nal.sh- submitspal2nal.shscript to pull corresponding nucleotides for all proteins usingpull_nucleotides.pyand prepares codon alignments using PAL2NAL../submit_hyphy.sh- prepares file inputs and submits scripts for HyPhy dN/dS methods -hyphy/BUSTED.sh,hyphy/aBSREL.shandhyphy/Contrast-FEL.sh.
cd selection/codon_optimisation
./pull_ribosomes.sh- extracts ribosomal protein encoding genes from Fusgr1 and submitsblast.shto run BLAST search against all strains in this study../submit_codon_optimisation.sh- submitscodon_optimisation.rscript to estimate various codon usage bias statistics and codon optimisation values.
Rscript stats_and_plots.r
Hill et al. (2022) Lifestyle transitions in fusarioid fungi are frequent and lack clear genomic signatures. Molecular Biology and Evolution 39(4):msac085. doi:10.1093/molbev/msac085.