ExplorATE (Explore Active Transposable Elements) is an R package for the exploration and identification of active transposons in RNA-seq data with or without reference genome. The package offers functions to manipulate the RepeatMasker output files, and allows to discriminate TEs-coding transcripts from those repeats that are co-transcribed with genes coding non-transposon proteins. Through a simple pipeline you can solve overlapping repeats that RepeatMasker cannot solve based on either higher score (HS), longer length (LE) or lower Kimura’s distances (LD). Additionally, the program offers optional refinement criteria for the annotation of TEs as a like-Wicker rule criterion based on the percentage of identity, the percentage of the repeat length respect to the transcript length, and a minimum length in base pairs. A decoy file and a transcriptome salmon-formated are created to be used for indexing and quantification with Salmon. Finally, a function is incorporated to import the transcripts quantification estimates into the R environment for subsequent differential expression analysis.
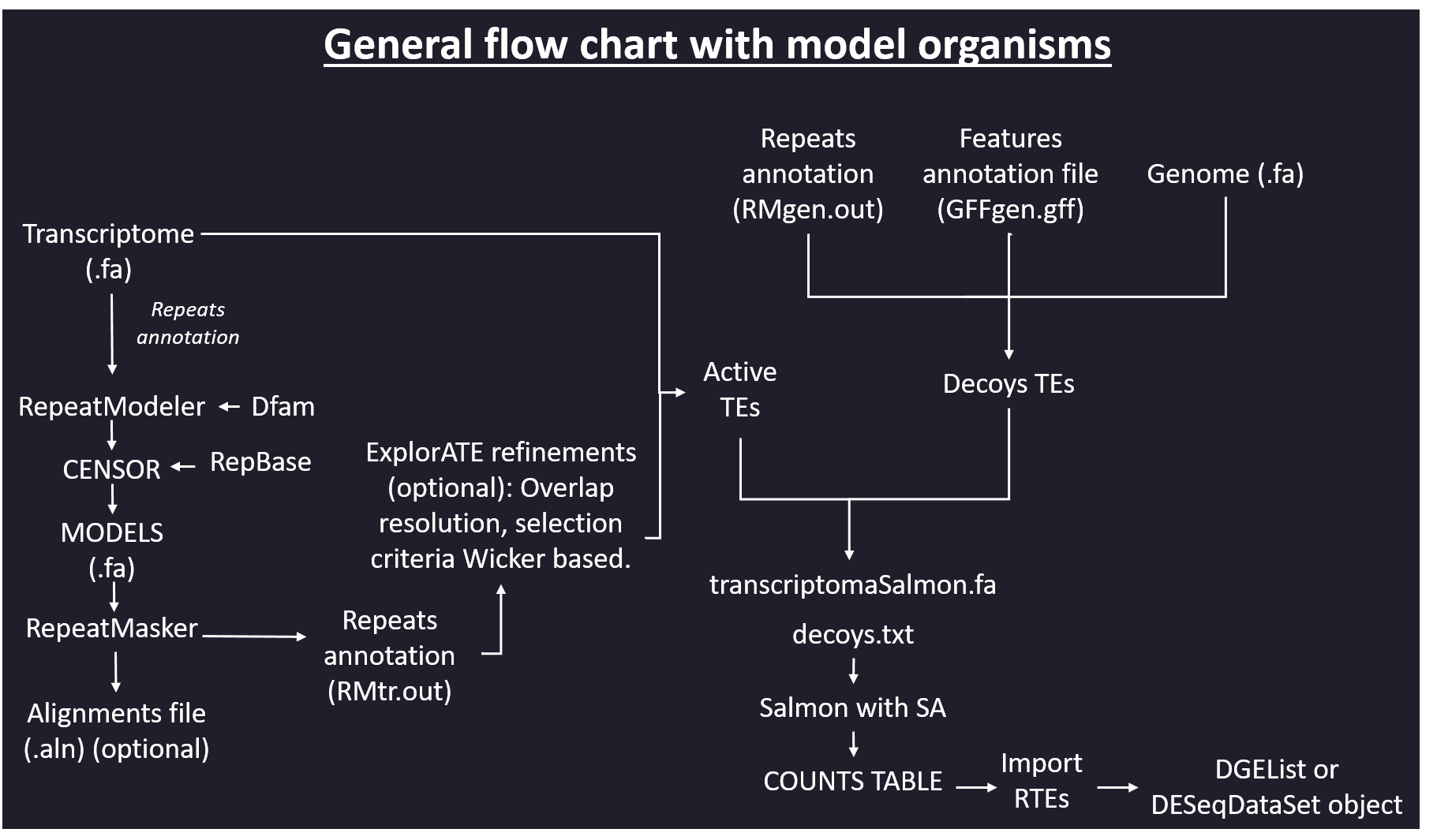
Flowchart with model organisms
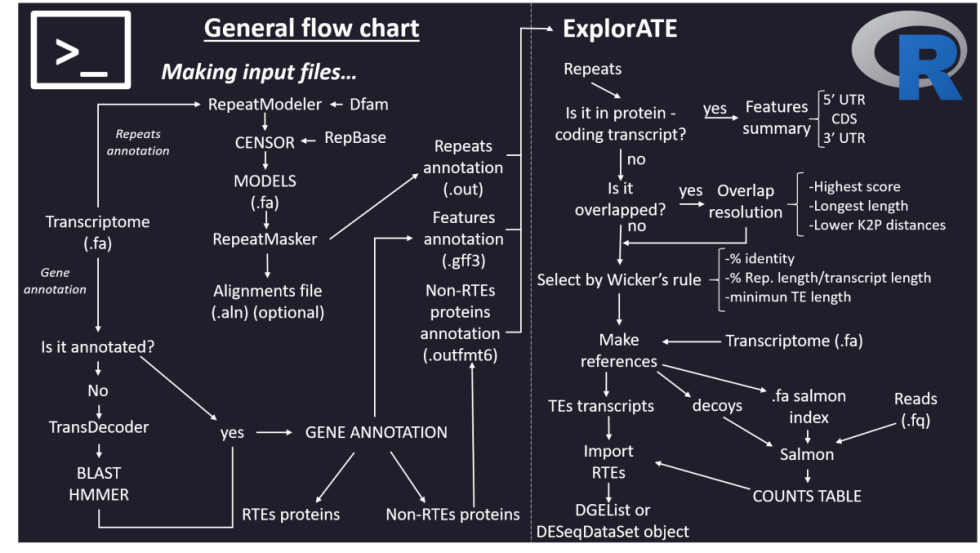
Flowchart with non-model organisms
ExplorATE requires bedtools to be installed on your system. If bedtools is not installed on your OS, please refer to this link and follow the instructions for your OS.
ExplorATE requires some previously installed R packages, select those packages that are not available in your environment:
install.packages(c("stringr","foreach","doParallel"))
install.packages(c("BiocManager","devtools"))
BiocManager::install(c("readr","GenomicRanges", "IRanges","csaw", "edgeR","SummarizedExperiment","DESeq2", "tximport"))
Install ExplorATE from GitHub
devtools::install_github("FemeniasM/ExplorATEproject")
Check the vignette and the user_guide