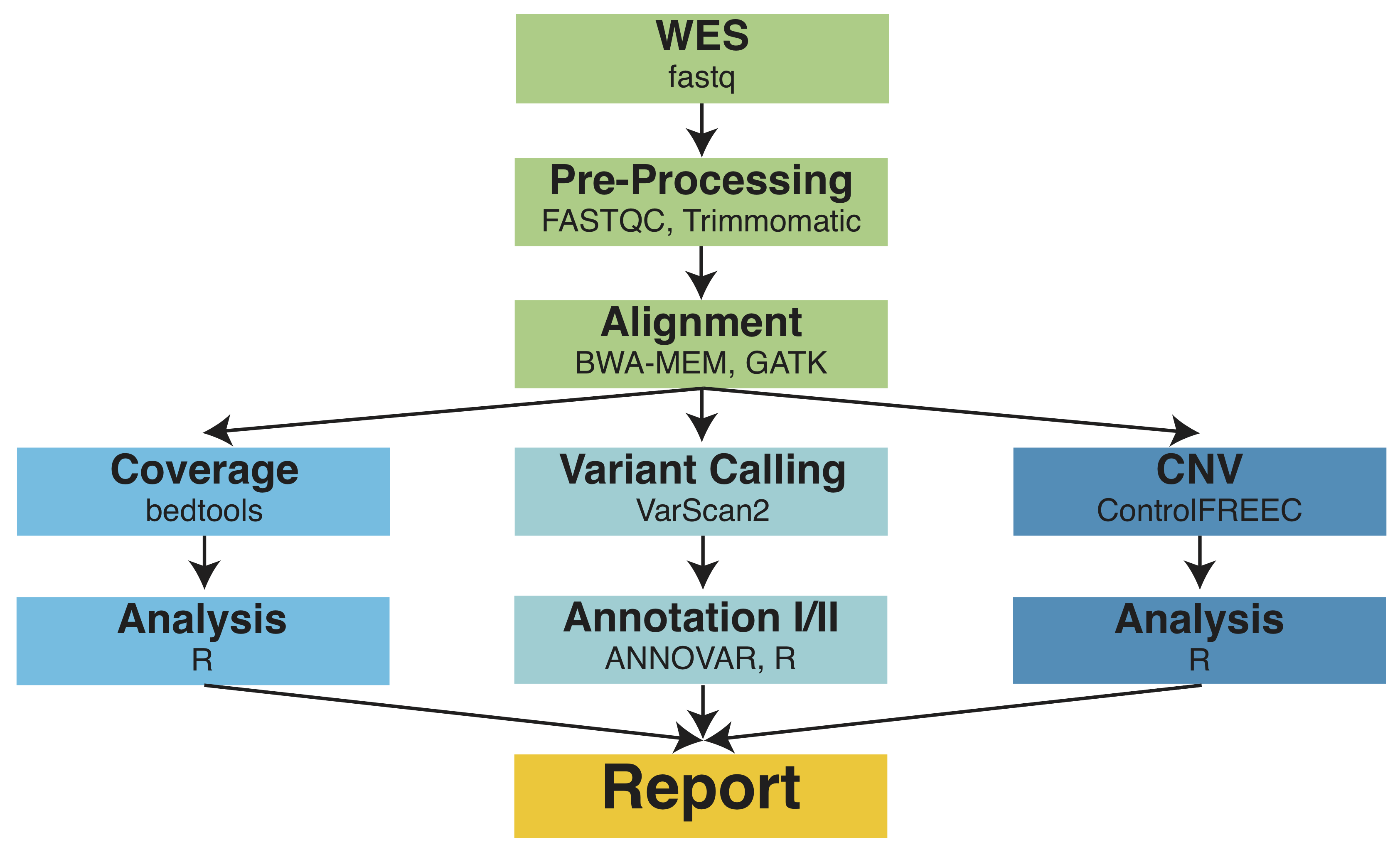
MIRACUM-Pipe incorporates tools for detecting single nucleotide variants (SNVs), insertions and deletions (InDels), loss of heterozygosity (LoH), copy number variations (CNVs) as well as quality statistics including the coverage on WES data. Various functional prediction and annotation databases are integrated to automatically annotate the identi-fied variants. The workflow is designed as a fully automated “one-click” solution from the raw sequencing files to the PDF report containing quality assessments, the identified and annotated variants (SNVs, InDels and LoH), copy number variations as well as a functional enrichment of the SNVs and CNVs respectively.
taken from Metzger P. et al. (submitted)
General: MIRACUM-Pipe is implemented using a bash script running all necessary programs and R scripts for further annotation and analysis of the results. Therefore, all tools and databases used have to be installed before running MIRACUM-Pipe. Although a default parameter set was established through extensive testing on various patients with different cancer entities and raw qualities, all parameters can be easily adjusted to the users’ needs as described later. The pipeline consists of three major parts, as depicted in Figure 1, namely (1) Alignment and Quality Control, (2) Analysis, Annotation and Interpretation (3) Composing of Results. The analysis part (2) is further divided in three subclasses, a) Coverage, b) Variant Calling and c) Copy Number Variations which were run in parallel.
Alignment and Quality Control: After quality control with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and adjustment with Trimmomatic (Bolger et al. 2014)), the burros-wheeler aligner (bwa-mem) (Li & Durbin 2010) is used to map the sequencing reads to the reference genome. To obtain more reliable sequence quality scores and alignment results we make use of the Genome Analysis Toolkit (GATK) (McKenna et al. 2010), specifically the tools BaseRecalibrator, RealignerTargetCreator, IndelRealigner and PrintReads, and the FixeMate function from picard-tools (http://broadinstitute.github.io/picard/).
Analysis, Annotation and Interpretation: For somatic variant calling, VarScan2 (Koboldt et al. 2013) is used. In parallel to the variant calling, the coverage is calculated with bedtools (Quinlan & Hall 2010) and the copy number variations are identified with Control-FREEC (Boeva et al. 2012). The identified variants are processed with ANNOVAR (Wang et al. 2010) and therefore annotated with the minor allele frequency (MAF) from gnomAD (Lek et al. 2016), SNP identifiers (dbSNP) (Sherry et al. 2001; Sherry et al. 1999), COSMIC IDs (Forbes et al. 2015), clinically interpreted from ClinVar (Landrum et al. 2014) and InterVar (Li & Wang 2017) and functionally annotated with dbNSFP (Liu et al. 2016). Additionally to the annotation with ANNOVAR, the variants are further annotated with the help of R scripts. First of all, the genes carrying a variant are classified in tumor suppressors or oncogenes according to OncoKB (Chakravarty et al. 2017). Cancer hotspot mutations are marked based on cancer-hotspots.org (Chang et al. 2016; Chang et al. 2017) and possible therapy options are identified from OncoKB (Chakravarty et al. 2017), TARGET (http://archive.broadinstitute.org/cancer/cga/target) and the drug-gene-interaction database (DGIdb) (Cotto et al. 2017).
Composing of Results: All identified and annotated variants and copy number variations are automatically composed to a PDF report. The report contains a general overview about the patient’s mutational burden, an overview about the number of SNVs and InDels, both separated for homozygous and heterozygous variants, and LoH. For the PDF report only exonic and variants leading to a protein change with a MAF < 0.1% and a variant allele frequency (VAF) > 10% are considered. The respected thresholds can be adjusted within the pipeline. The variants are further categorized in tumor suppressors, oncogenes and cancer hotspots. The latter are all listed in a table together with the top somatic variants according to VAF. For better understanding of the biological processes affected by the variants, a functional enrichment is calculated on Gene Ontology (GO) terms (Ashburner et al. 2000; Blake et al. 2015), ConsensusPathDB (Kamburov et al. 2012; Kamburov et al. 2013) and Reactome (Fabregat et al. 2018). The signaling pathways with the highest significance are reported. Additionally, the variants are checked against five cancers associated signaling pathways, namely PI3K-AKT-mTOR, RAF-MEK-ERK, DNA Damage Response, Cell Cycle and Tyrosine Kinases that play a role for cancer processes. The pathways were taken from Qiagen (https://www.qiagen.com/). Further, the COSMIC mutational signatures, e.g. BRCAness signature (AC3, DNA damage) that gives insight for therapeutical options like PARP-Inhibitors, according to (Alexandrov et al. 2013) are calculated. The copy number variations are visualized and explicitly reported for tumor suppressors and oncogenes. For better insight in the altered processes by the chromosomal instabilities a functional enrichment based on GO terms, ConsensusPathDB and Reactome is performed. Last but not least, all used tools and databases, including version informations, are reported.
Requiered environment, tools and databases.
-
Environment
- Torque Scheduler (qsub command needed)
- Reference genome hg19 inclunding index files for bwa
-
Software
- java (1.8.0_152)
- FASTQC (v0.11.5)
- Trimmomatic (0.38)
- bwa (0.7.17)
- bam-readcount (0.8.0-unstable-6-963acab)
- samtools (1.9)
- GATK (version 3.8-1-0-gf15c1c3ef)
- picard-tools (2.18.15)
- bedtools (v2.27.1)
- VarScan2 (2.4.3)
- ANNOVAR ($Date: 2018-04-16 00:47:49 -0400)
- snpEff (4.3t)
- Control-FREEC (11.0)
- R (3.4.1)
- LaTeX
-
Databases from ANNOVAR
- refGene
- gnomAD_exome
- exac03
- esp6500siv2_ea
- EUR.sites.2015_08
- avsnp150
- clinvar_20180603
- cadd13
- intervar_20180118
- dbnsfp35a
- cosmic86
-
R package
- OmicCircos
- foreach
- doMC
- rtracklayer
- openxlsx
- org.Hs.eg.db
- GOstats
- Homo.sapiens
- biomaRt
- RColorBrewer
- stringr
- Rsamtools
- gtrellis
- circlize
- ComplexHeatmap
- YAPSA
- SomaticSignatures
- VariantAnnotation
- BSgenome.Hsapiens.UCSC.hg19
- TxDb.Hsapiens.UCSC.hg19.knownGene
- ReactomePA
- knitr
- docstring
-
Additional Files
- CaptureRegions, e.g. Agilent SureSelect V5UTR according to the used CaptureKit
- dbSNP vcf file corresbonding to the one used from ANNOVAR, e.g. snp150hg19.vcf.gz
- FLAG genes (included, flag_genes.txt) (Shyr et al., 2014)
- CancerGenes from OncoKB (needed)
- Cancer hotspots from cancerhotspots.org (needed)
- RVIS score (included, RVIS_score.txt) (Petrovski S. et al., 2013)
- TARGET db (included, TARGET_db.txt) (https://software.broadinstitute.org/cancer/cga/target)
- DGIdb from www.dgidb.org (needed)
- Actionable alterations from OncoKB (needed)
- Condel score from FannsDB (needed)
- captured genes according to the used CaptureKit (Targets.txt supplied for Agilent SureSelect)
- Cancer associated pathways (included, MTB_GeneSets.RData)
- Consensus Pathways (included, Consensus.RData)
- Reactome Pathways (included, Reactome.RData)
- GO Pathways (included, GOGeneSets.RData)
- HallmarksOfCancer (needed)
If available, the used versions are noted.
MIRACUM-Pipe is currently designed to work with the following capture kits:
* Agilent SureSelect V6
* Agilent SureSelect V5UTR
But it can be easily adjusted to any other capture kit as long as the captureRegions .bed file is available, the covered genes and the region covered in Mega-bases (MB) is known. If a different Capture Kit is used the covered regions in MB has to be added to the RScript filtering_tools.R in the function tumbu starting in line 3. Additionally the covered genes have to be stored in a plain text file named Targets.txt and the captureRegions.bed file needs to be placed in the used annotation folder.
dbSNP from https://www.ncbi.nlm.nih.gov/projects/SNP/
- Actionable Alterations (OncoKB) from http://oncokb.org/#/dataAccess -> allActionableVariants.txt
- CancerGenes (OncoKB) from https://github.com/oncokb/oncokb-public/blob/master/data/v1.15/CancerGenesList.txt -> CancerGenesList.txt
- Cancer Hotspots from http://www.cancerhotspots.org/#/download (hotspots_v2.xls) -> save the two excel sheets into separate tab-separated text files -> hotspots_v2.txt and hotspots_v2_indel.txt
- DGIdb from http://www.dgidb.org/downloads (Interactions TSV) -> DGIdb_interactions.tsv
- Condel from http://bbglab.irbbarcelona.org/fannsdb/home (fannsdb.tsv.gz + fannsdb.tsv.gz.tbi)
- HallmarksOfCancer from http://software.broadinstitute.org/gsea/msigdb/index.jsp converted to a RData file -> hallmarksOfCancer_GeneSets.Rdata
All those databases have to be saved in the Databases folder.
After all tools and databases are installed and work properly, MIRACUM-Pipe can be used after adjusting three script to the environment of the working system. The scripts createSet.sh and make_alignment_VC_CNV.sh have only to be adjusted once for the working environment. The thrid script run.sh is needed for every sample and contains the sample ID and the path to the raw data.
The script createSet.sh contains all the code which is neccessary to create the single job submissions for alignment, variant calling, copy number calling and report generation. Therefore the path to the output folders, namely WES and Analysis, and the directory containing the later folders together with the bash scripts have to be adjusted to meet local conditions in lines 27ff. The WES folder contains all results from quality control, alignment, variant calling and the output from ANNOVAR. In the Analysis folder the final tables from further variant annotations, all analysis performed in R, e.g. GeneSet Enrichemnt Analysis, and the report are stored.
##################################################################################################################
## Parameters which need to be adjusted to the local environment
mtb="/path/to/output/folder/${case}_${num}" # path to output folder, subfolder with type of analysis and ID is automatically created
wes="${mtb}/WES" # subfolder containing the alignemnt, coverage, copy number variation and variant calling results
ana="${mtb}/Analysis" # subfolder containing PDF Report, annotated copy number variations and annotated variants
##################################################################################################################
The script make_alignment_VC_CNV.sh contains the actual function calls to perfom quality control, alignemnt, variant calling, copy number calling, annotation and report generation. It has to be adjusted to the local conditions starting in line 14ff. Our set of parametes is extensivley tested on various patients with different cancer entities and raw qualities and delivers reliable and consitent results. But nervertheless it might need adjustments from time to time which can be easily achieved in this script starting at line 176ff. Here all parameters can be adjusted, e.g. VAF cutoff.
##################################################################################################################
#### Parameters which have to be adjusted accoridng the the environment or the users needs
## General
homedata="/path/to/data" # folder contatining the raw data (.fastq files)
annot="/path/to/annotation" # folder containing annotation files like captureRegions.bed
motherpath="/path/to/output"
mtb="${motherpath}/${case}_${num}" # folder containing output
wes="${mtb}/WES"
ana="${mtb}/Analysis"
RscriptPath="${motherpath}/RScripts"
DatabasePath="${motherpath}/Databases"
tempdir="/scratch" # temporary folder
## Genome
GENOME="/path/to/ref/genome/including/index/hg19.fa"
Chromosomes="/path/to/ref/genome/chromosomes"
ChromoLength="/path/to/ref/chromosomes/length/hg19_chr.len"
## SureSelect (Capture Kit)
CaptureRegions="${annot}/Agilent/SureSelectV5UTR/V5UTR.bed"
## dbSNP vcf File
dbSNPvcf="/path/to/dbSNP/dbSNP/snp150hg19.vcf.gz "
### Software
## Parameters
## General
# Cores to use
nCore=12
minBaseQual="28"
minVAF="0.10"
# VarScan somatic
minCoverage="8"
TumorPurity="0.5"
minFreqForHom="0.75" # VAF to call homozygote
# VarScan fpfilter
minVarCount="4"
# ANNOVAR Databases
protocol='refGene,gnomad_exome,exac03,esp6500siv2_ea,EUR.sites.2015_08,avsnp150,clinvar_20180603,intervar_20180118,dbnsfp35a,cosmic86_coding,cosmic86_noncoding'
argop='g,f,f,f,f,f,f,f,f,f,f'
## Tools and paths
# Paths
soft="/path/to/tools/software" # folder containing all used tools
java="${soft}/bin/java -Djava.io.tmpdir=${tempdir} " # path to java
# Pre-Processing
FASTQC="${soft}/FastQC/fastqc -t ${nCore} --extract "
TRIM="${java} -Xmx150g -jar ${soft}/Trimmomatic-0.36/trimmomatic-0.36.jar PE -threads ${nCore} -phred33 "
CUT="cut -f1,2,3"
# Alignment
BWAMEM="${soft}/bin/bwa mem -M "
# BAM-Readcount
BamReadcount="${soft}/bin/bam-readcount -q 1 -b 20 -w 1 -f ${GENOME} "
# SAMTOOLS
SAMTOOLS="${soft}/bin/samtools" # path to samtools
SAMVIEW="${SAMTOOLS} view -@ ${nCore} "
SAMSORT="${SAMTOOLS} sort -@ ${nCore} "
SAMRMDUP="${SAMTOOLS} rmdup "
SAMINDEX="${SAMTOOLS} index "
MPILEUP="${SAMTOOLS} mpileup -B -C 50 -f ${GENOME} -q 1 --min-BQ ${minBaseQual}"
STATS="${SAMTOOLS} stats "
# GATK
GATK="${soft}/bin/gatk"
RealignerTargetCreator="${GATK} -T RealignerTargetCreator -R ${GENOME} -nt ${nCore} "
IndelRealigner="${GATK} -R ${GENOME} -T IndelRealigner "
BaseRecalibrator="${GATK} -T BaseRecalibrator -l INFO -R ${GENOME} -knownSites ${dbSNPvcf} -nct ${nCore} "
PrintReads="${GATK} -T PrintReads -R ${GENOME} -nct ${nCore} "
# PICARD
FixMate="${soft}/bin/picard FixMateInformation "
# VARSCAN
VarScan="${soft}/anaconda2/bin/varscan"
SOMATIC="${VarScan} somatic"
PROCESSSOMATIC="${VarScan} processSomatic"
# ANNOVAR
CONVERT2ANNOVAR2="${soft}/annovar/convert2annovar.pl --format vcf4old --outfile "
CONVERT2ANNOVAR3="${soft}/annovar/convert2annovar.pl --format vcf4old --includeinfo --comment --outfile "
CONVERT2ANNOVAR="${soft}/annovar/convert2annovar.pl --format vcf4 --includeinfo --comment --withzyg --outfile "
TABLEANNOVAR="${soft}/annovar/table_annovar.pl"
# COVERAGE
COVERAGE="${soft}/bedtools2/bin/bedtools coverage -hist -g ${GENOME}.fai -sorted "
# SNPEFF
SNPEFF="${java} -Xmx150g -jar ${soft}/snpEff/snpEff.jar GRCh37.75 -c ${soft}/snpEff/snpEff.config -canon -v"
# ControlFREEC
freec="${soft}/bin/freec "
gemMappabilityFile="${soft}/FREEC-11.0/mappability/out100m2_hg19.gem"
# R
Rscript="${soft}/bin/Rscript"
The run.sh script needs to be adjusted for every sample / patient. It calls the createSet script to create the separate jobs for alignment, variant calling, copy number calling, etc. Therefore, the patient ID, analysis type, path to the raw files and the filenames have to be given as well as the gender of the patient. The analysis type could be either somatic or somaticGermline. During somatic the germline sample is only used to identify somatic variations which were further processed and annotated. For somaticGermline, the germline variants were called, further processed and annotated as well.
# createSet.sh and make_alignment_VC_CNV.sh need to be in the base directory
# first germline (GD), then tumor (TD)
./createSet.sh somaticGermline ID 'folder/containing/germline' 'folder/containing/tumor' filename_germline_without_file_extension filename_tumor_without_file_extension XY
cd somaticGermline_ID
./run_jobs.sh > out &
For the databases contained in ANNOVAR updating or installing new databases is very simple following the descriptions on the homepage of ANNOVAR. If the additional database is installed it has to be added in line 117 and 118 in the make_alignment_VC_CNV.sh script. Adding a database currently not supported by ANNOVAR is also possible but one has to add a few lines and the actual database to the corresponding part in the filtering.R script. See the documentation of the R scripts.
All neccessary informations about the included R-scripts can be found with the ?functionName function in R. The predefined report in Report.rnw contains in line 45 the author name which should be adjusted to the person generating the report.
For testing MIRACUM-Pipe the public available dataset from Texas (http://txcrb.org/data.html and https://www.nature.com/articles/sdata201610) can be used. We provide the PDF reports for the samples TCRBOA6 and TCRBOA7. Both samples were run with the default parameters.
- Patrick Metzger
- Maria Hess
- Victor Jaravine
- Jochen Hochrein
- Martin Boeker
- Hauke Busch
- Geoffroy Andrieux
- Melanie Börries
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
We thank
- The Molecular Tumor Board Freburg Team
- The whole MIRACUM consortia
- The German Ministry of Education and Research (BMBF) for funding
- The developers from Control-FREEC for the code on CNV significance
If you use this work please cite Metzger P. et al., (submitted)
Alexandrov, L.B. et al., 2013. Signatures of mutational processes in human cancer. Nature, 500, 415–421.
Ashburner, M. et al., 2000. Gene Ontology: Tool for The Unification of Biology. Nature Genetics, 25, 25–29.
Blake, J.A. et al., 2015. Gene ontology consortium: Going forward. Nucleic Acids Research, 43, D1049–D1056.
Boeva, V. et al., 2012. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics, 28, 423–425.
Chakravarty, D. et al., 2017. OncoKB: a precision oncology knowledgebase. Precision Oncology.
Chang, M.T. et al., 2017. Accelerating discovery of functional mutant alleles in cancer. Cancer Discovery, 170321.
Chang, M.T. et al., 2016. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational speci-ficity. Nature Biotechnology, 34, 155–163.
Cotto, K.C. et al., 2017. DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Research, 46, 1068–1073.
Fabregat, A. et al., 2018. The Reactome Pathway Knowledgebase. Nucleic Acids Research, 46, D649–D655.
Forbes, S.A. et al., 2015. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic acids research, 43, D805-11.
Hoefflin, R. et al., 2018. Personalized Clinical Decision Making Through Implementation of a Molecular Tumor Board: A German Single-Center Experience. JCO Precision Oncology, 2, 1–16.
Kamburov, A. et al., 2012. ConsensusPathDB : assembling a more complete picture of cell biology, 2797.
Kamburov, A. et al., 2013. The ConsensusPathDB interaction database: 2013 update. Nucleic acids research, 41, D793-800.
Koboldt, D.C., Larson, D.E. & Wilson, R.K., 2013. Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. In Current Protocols in Bioinformatics. Hoboken, NJ, USA: John Wiley & Sons, Inc., 15.4.1-15.4.17.
Landrum, M.J. et al., 2014. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic acids research, 42, D980-5.
Lek, M. et al., 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291.
Li, Q. & Wang, K., 2017. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. American Journal of Human Genetics, 100, 267–280.
Liu, X. et al., 2016. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Human Mutation, 37, 235–241.
Petrovski S. et al., PLoS Genet, 2013
Quinlan, A.R. & Hall, I.M., 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842.
Sherry, S.T. et al., 2001. dbSNP: the NCBI database of genetic variation. Nucleic acids research, 29, 308–11.
Sherry, S.T., Ward, M. & Sirotkin, K., 1999. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome research, 9, 677–9.
Shyr et al., BMC Medical Genomics 2014
Wang, K., Li, M. & Hakonarson, H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research, 38(16), p.e164.