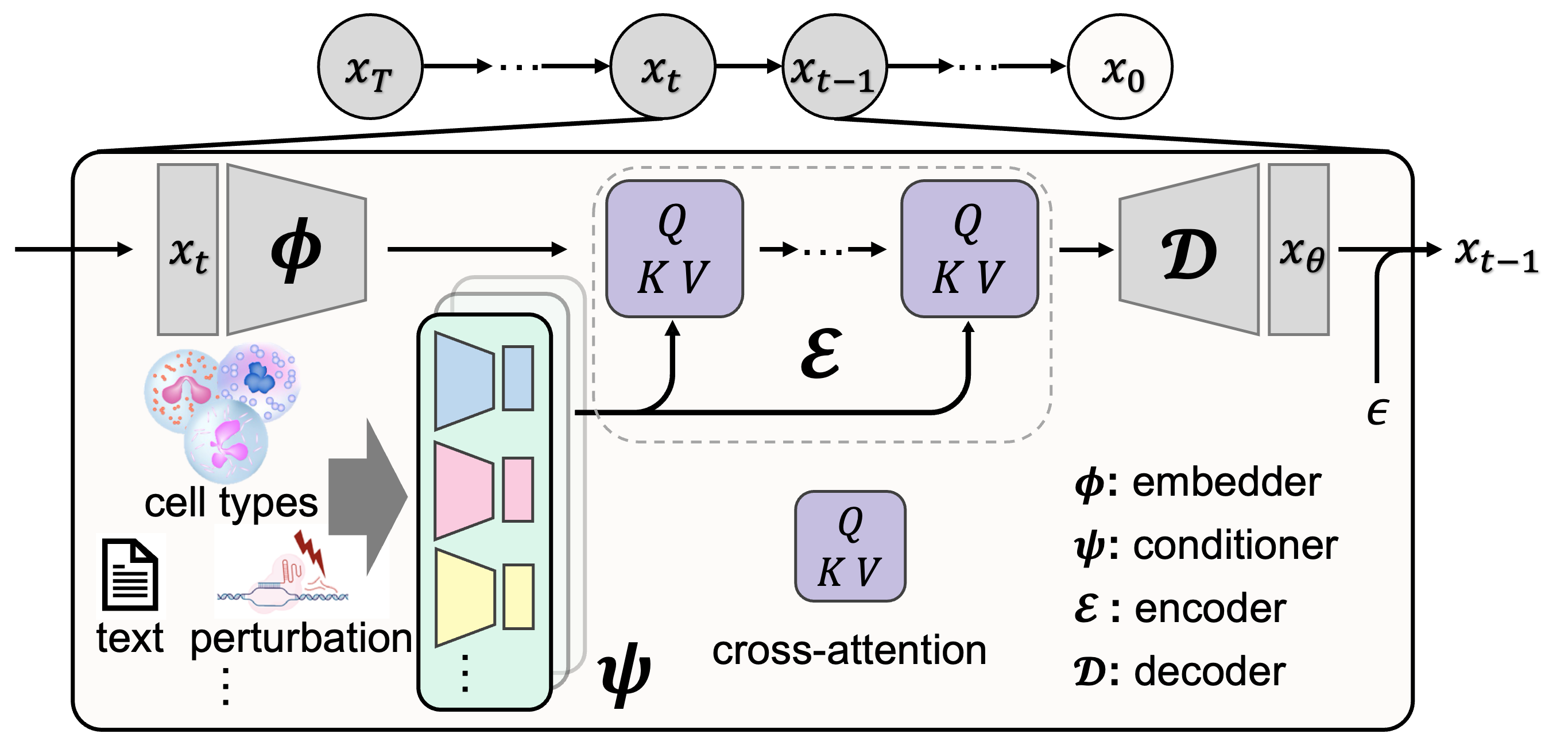
This is the official codebase for A General Single-Cell Analysis Framework via Conditional Diffusion Generative Models.
scDiff achieves competitive performance with state-of-the-art models in various benchmarking tasks. By incorporating prior knowledge with large language models and graph neural networks, scDiff shows outstanding few-shot and zero-shot results.
conda create -n scdiff python=3.9 -y && conda activate scdiff
# PyTorch
conda install pytorch==2.0.1 torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia -y
# PyG
conda install pyg==2.3.1 -c pyg
# Other dependencies
pip install -r requirements.txt
# scDiff
pip install -e .
conda clean --all -y# Install gdown for downloading data and pre-trained models
pip install gdown
gdown https://drive.google.com/file/d/1Dbf1aX-OZ8FOGVEi4PhKiDRcvBOrnyAI/view?usp=sharing --fuzzy
tar -xzvf reproducibility.tar.gz && mv reproduce/ data/### Annotation experiments
SEED=10 bash run/run.sh annotation
### Imputation experiments
SEED=10 bash run/run.sh denoising
### Perturbation experiments
SEED=10 bash run/run.sh perturbation
### Gene perturbation experiments
SEED=10 bash run/run.sh genepert
### One-shot annotation experiments
SEED=10 TOPK=3 bash run/oneshot.sh