Note: This code repository is no longer maintained. The updated code can be found here.
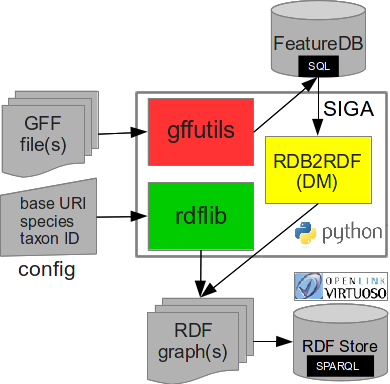
SIGA.py is a command-line tool to generate Semantically Interoperable Genome Annotations from GFF files according to the Resource Description Framework (RDF) specification.
- process multiple input files in GFF (versions 2 and 3)
- genome annotations (features) stored in SQLite database and serialized as RDF graph(s) in plain text formats:
- supported feature (keys) types: genome, chromosome, gene, prim_transcript, mRNA, CDS, exon, intron, five_prime_UTR, three_prime_UTR, polyA_site, polyA_sequence
- supported feature relations (SO(FA) properties): has_part and its inverse part_of, transcribed_to, genome_of
- sequence feature locations described by FALDO
- parent-child feature relationships checked for referential integrity
Python 2.7
docopt 0.6.2
RDFLib 4.2.2
gffutils (https://github.com/arnikz/gffutils)
RDF store (e.g. Virtuoso or Berkeley DB) to ingest and query data using SPARQL
Install and activate virtualenv
virtualenv sigaenv
source sigaenv/bin/activate
Use requirements.txt from repository to update the virtual env with the necessary packages:
pip install -r requirements.txt
Example data
The sample genome annotations are located in the examples folder
cd examples
Alternatively, one can download the latest genome annotations for tomato (ITAG v2.4)
wget ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/ITAG2.4_release/ITAG2.4_gene_models.gff3
or potato (PGSC v4.03)
wget http://solanaceae.plantbiology.msu.edu/data/PGSC_DM_V403_genes.gff.zip
Example usage
cd src
Two-steps process to serialize triples in RDF Turtle (default):
-
GFF to DB
python SIGA.py db -rV ../examples/ITAG2.4_gene_models.gff3 -
DB to RDF
python SIGA.py rdf \ -b https://solgenomics.net/ \ -c http://orcid.org/0000-0003-1711-7961 \ -s ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/ITAG2.4_release/ITAG2.4_gene_models.gff3 \ -n "Solanum lycopersicum" \ -t 4081 ITAG2.4_gene_models.dbor with a config.ini file
python SIGA.py rdf -C config.ini ../examples/ITAG2.4_gene_models.db
Summary of input/output files:
ITAG2.4_gene_models.gff3 # GFF file
ITAG2.4_gene_models.db # SQLite database
ITAG2.4_gene_models.ttl # RDF file in Turtle
Import RDF graph into Virtuoso RDF Quad Store
See the documentation on bulk data loading.
Edit virtuoso.ini config file by adding /mydir/ to DirsAllowed.
Connect to db server as dba user:
isql 1111 dba dba
Delete (old) RDF graph if necessary:
SPARQL CLEAR GRAPH <https://solgenomics.net/genome/Solanum_lycopersicum> ;
Delete any previously registered data files:
DELETE FROM DB.DBA.load_list ;
Register data file(s):
ld_dir('/mydir/', 'ITAG2.4_gene_models.ttl', 'https://solgenomics.net/genome/Solanum_lycopersicum') ;
List registered data file(s):
SELECT * FROM DB.DBA.load_list ;
Bulk data loading:
rdf_loader_run() ;
Re-index triples for full-text search:
DB.DBA.VT_INC_INDEX_DB_DBA_RDF_OBJ() ;
Note: For loading a single data file one could use the following command:
SPARQL LOAD "file:///mydir/ITAG2.4_gene_models.ttl" INTO "https://solgenomics.net/genome/Solanum_lycopersicum" ;
However, this approach results in additional triples (generated by Virtuoso) which are not present in the input file.
Count imported triples:
SPARQL
SELECT COUNT(*)
FROM <https://solgenomics.net/genome/Solanum_lycopersicum>
WHERE { ?s ?p ?o } ;
Alternatively, persist RDF graph in Berkeley DB using the Redland RDF processor
rdfproc ITAG2.4_gene_models parse ITAG2.4_gene_models.ttl turtle
rdfproc tomato_QTLs serialize turtle
Please, refer to SIGA.py in scientific publications by this persistent identifier:
The software is released under Apache License 2.0 licence.