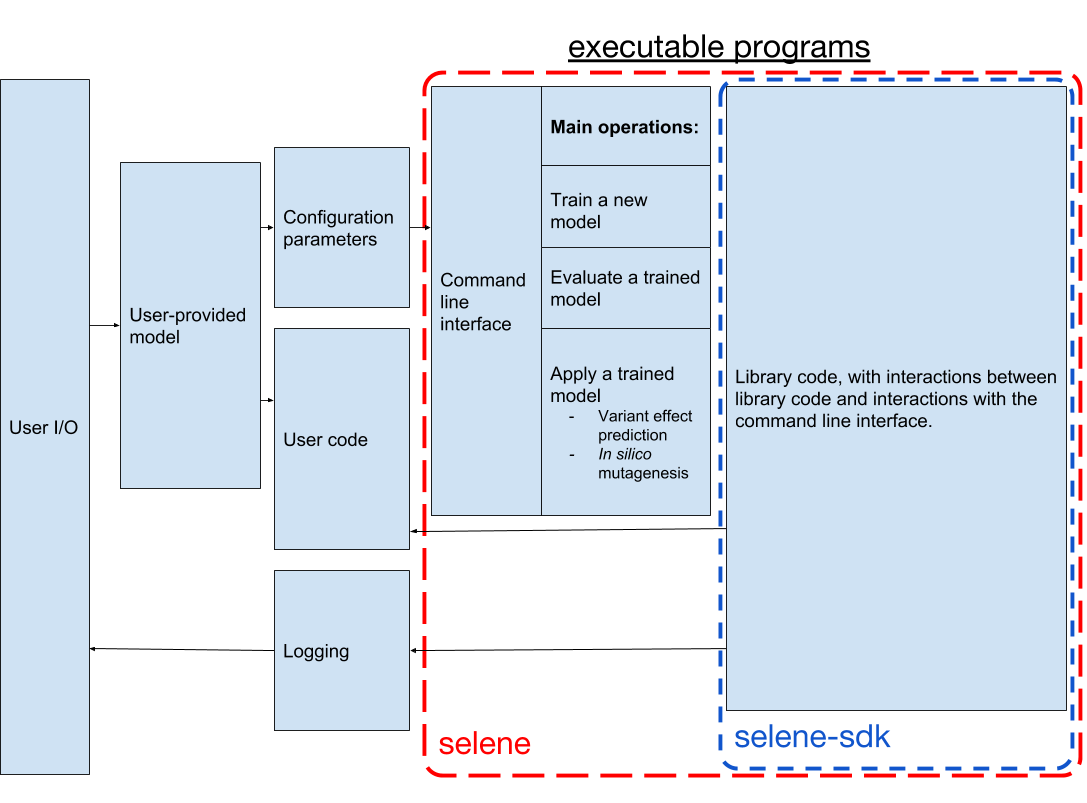
You have found Selene, a Python library and command line interface for training deep neural networks from biological sequence data such as genomes.
Selene is a Python 3+ package. We recommend using it with Python 3.6 or above. Package installation should only take a few minutes (less than 10 minutes, typically ~2-3 minutes) with any of these methods (pip, conda, source).
Installing selene with Anaconda (for Linux):
conda install -c bioconda selene-sdkpip install selene-sdkFirst, download the latest commits from the source repository (or download the latest tagged version of Selene for a stable release):
git clone https://github.com/FunctionLab/selene.git
The setup.py script requires NumPy. Please make sure you have this already installed.
If you plan on working in the selene repository directly, we recommend setting up a conda environment using selene-cpu.yml or selene-gpu.yml (if CUDA is enabled on your machine) and activating it.
These environment YAML files list specific versions of package dependencies that we have used in the past to test Selene.
Selene contains some Cython files. You can build these by running
python setup.py build_ext --inplaceOtherwise, if you would like to locally install Selene, you can run
python setup.py installPlease install docopt before running the command-line script selene_cli.py provided in the repository.
Selene is composed of a command-line interface and an API (the selene-sdk Python package).
Users supply their data, model architecture, and configuration parameters, and Selene runs the user-specified operations (training, evaluation, prediction) for that sequence-based model.
For a more detailed overview of the components in the Selene software development kit (SDK), please consult the page here.
The documentation for Selene is available here.
In general, we recommend that the manuscript case studies and the tutorials be run on a machine with a GPU. All examples take significantly longer when run on a CPU machine.
Tutorials for Selene are available here.
It is possible to run the tutorials (Jupyter notebook examples) on a standard CPU machine--you should not expect to fully finish running the training examples unless you can run them for more than 2-3 days, but they can all be run to completion on CPU in a couple of days. You can also change the training parameters (e.g. total number of steps) so that they complete in a much faster amount of time.
The non-training examples (variant effect prediction, in silico mutagenesis) can be run fairly quickly (variant effect prediction might take 20-30 minutes, in silico mutagenesis in 10-15 minutes).
Please see the README in the tutorials directory for links and descriptions to the specific tutorials.
The code to reproduce case studies in the manuscript is available here.
Each case has its own directory and README describing how to run these cases. We recommend consulting the step-by-step breakdown of each case study that we provide in the methods section of the manuscript as well.
The manuscript examples were only tested on GPU. Our GPU (NVIDIA Tesla V100) time estimates:
- Case study 1 finishes in about 1 day on a GPU node.
- Case study 2 takes 6-7 days to run training (distributed the work across 4 v100s).
- Case study 3 (variant effect prediction) takes about 1 day to run.