install.packages("MERO")Perform missing value imputation for biological data using the random forest algorithm, the imputation aim to keep the original mean and standard deviation consistent after imputation.
For the documentation see: MERO Documentation.
- link to package on CRAN: MERO
Example
library(MERO)
library(missForest)
#Load a sample data
data(iris)
summary(iris)
## The data contains four continuous and one categorical variable.
## Artificially produce missing values using the 'prodNA' function:
iris.mis <- prodNA(iris, noNA = 0.2)
summary(iris.mis)
#Impute the missing data using random forest
#Nsets is the number of data sets to be imputed/ the number of runs or simulations
#ntree is the number of trees for random forest
Imp.data <- MERO(Data = iris.mis[,1:4], ntree = 100, Nsets = 5)
#Select the best data set which mean and standard deviation are very close to the original mean and standard deviation of the input data
Best.hit <- EvalImp(Originaldata = iris.mis[,1:4], ImputedSets = Imp.data[[1]],
Imputed.mean = Imp.data[[2]], Imp.data[[3]])
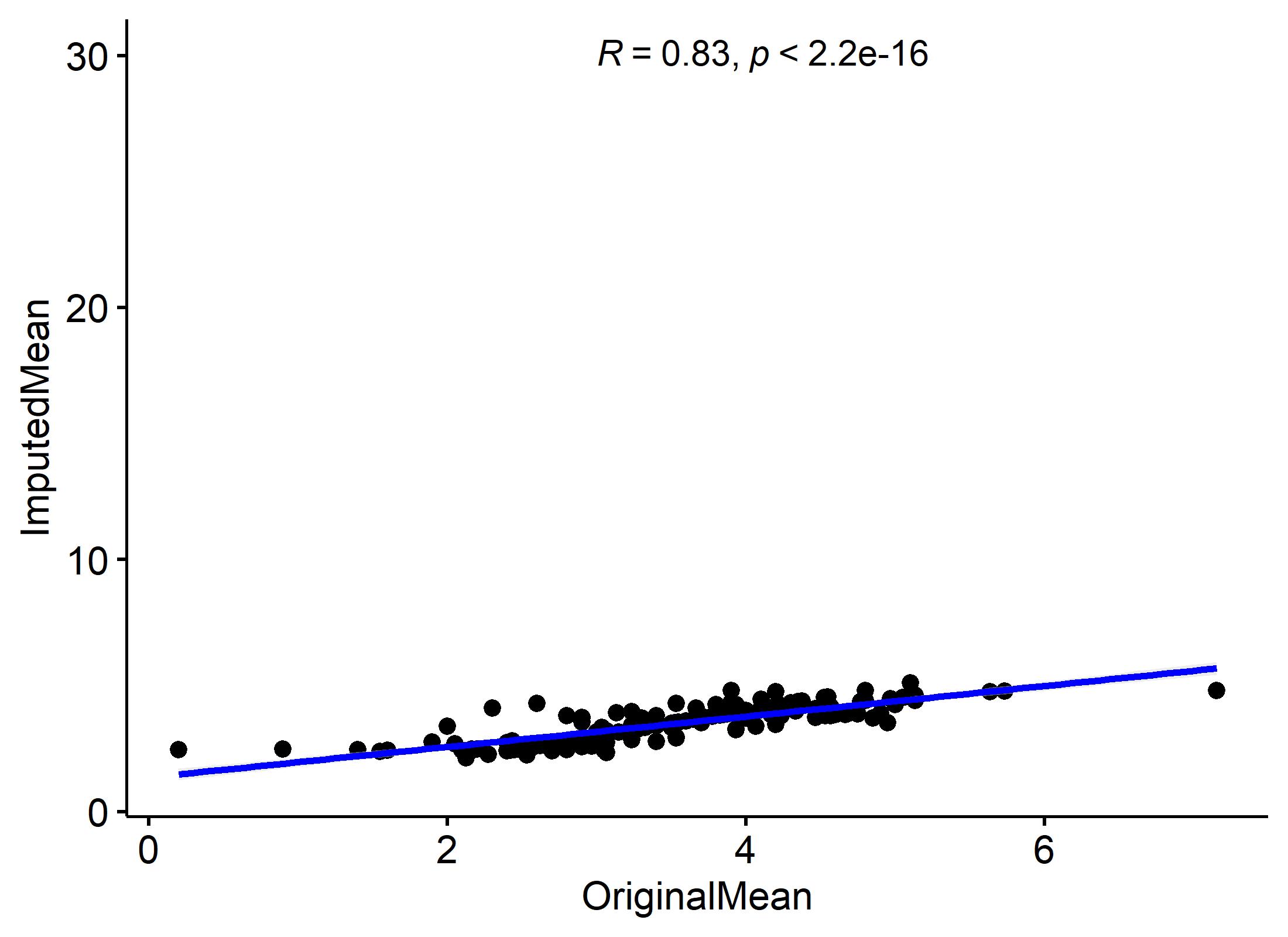
#Visualize the correlation between the original means and the imputed means of the data sets
PlotCorrelateMean(Best.hit[[2]],Best.hit[[3]])For bugs and suggestions, the most effective way is by raising an issue on the github issue tracker. Github allows you to classify your issues so that we know if it is a bug report, feature request or feedback to the authors.