There are some useful scripts.
A R script for transforming vcf file to SNPBinner input file. The heterozygosity and missing rate will be calculated for output SNP marker.
Rscript ./vcf2snpbinner.R -husage: reform.R [--] [--help] [--opts OPTS] [--input INPUT] [--out OUT]
[--parent1 PARENT1] [--parent2 PARENT2] [--minDP_p1 MINDP_P1]
[--minDP_p2 MINDP_P2]
a program for converting vcf to table of snpbinner. genotype same as
parent_1 is designated 'a', genotype same as parent_2 is designated
'b', heterozygous genotype is designated 'h', missing genotype is
designated '-'
flags:
-h, --help show this help message and exit
optional arguments:
-x, --opts RDS file containing argument values
-i, --input vcf or vcf.gz file containing two parents and progeny
lines
-o, --out output file prefix
-p, --parent1 name of parent_1
--parent2 name of parent_2
-m, --minDP_p1 Minimum depth of parent_1 [default: 5]
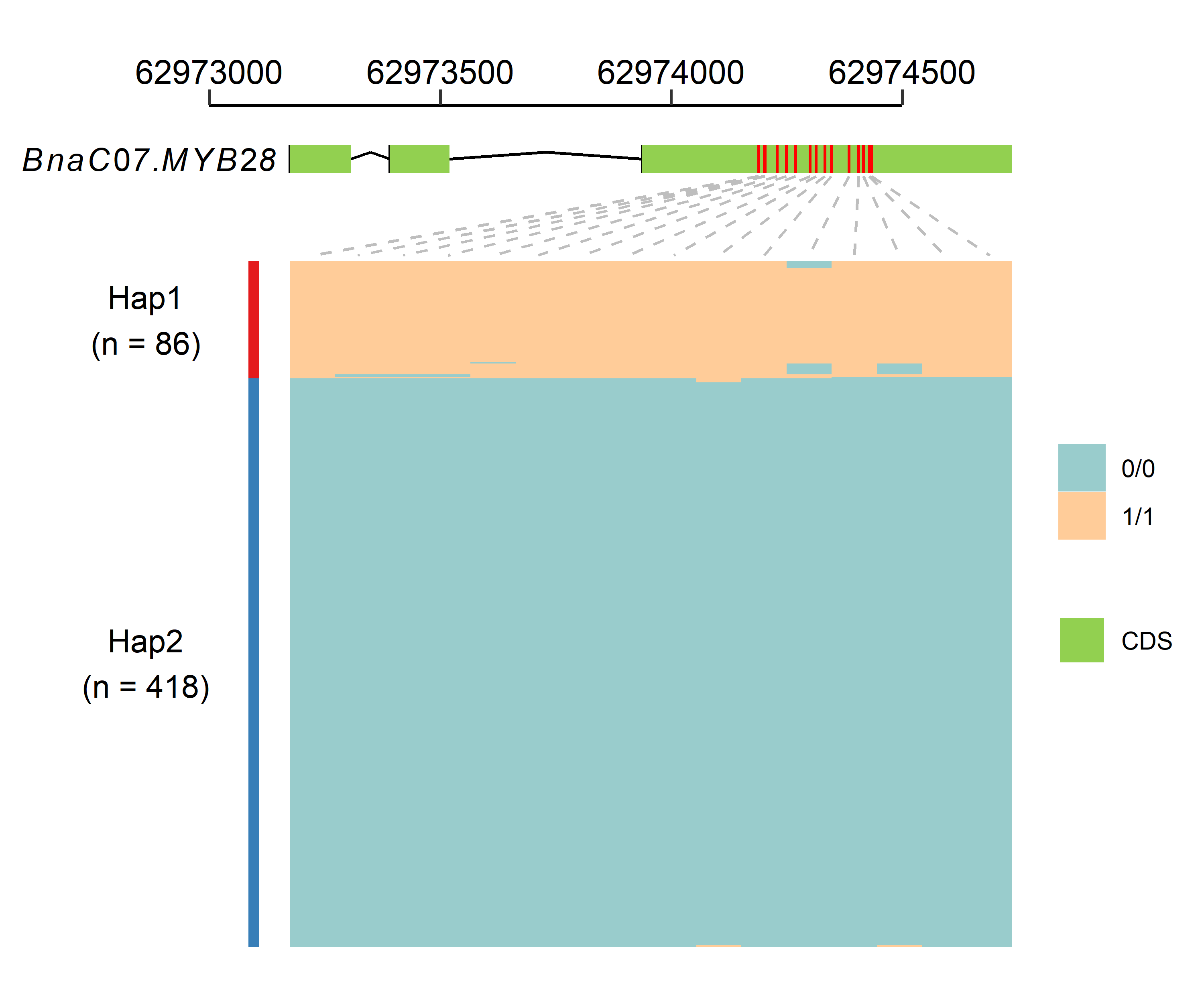
--minDP_p2 Minimum depth of parent_2 [default: 5]A R script for drawing gene structure and the variation of this gene in a population. A gtf file containing target gene, a vcf file containing variation of this gene and phenotype data is needed. Hierarchical clustering algorithm was adopted to distinguish different haplotype, the number of haplotype can be designated according clusting result. Some polymorphism may exsit within samples belonging to the same haplotype, you can divided them into different haplotypes by setting more haplotypes.
A perl script for ePCR. Input is tsv (tab-separated values) file containing three columns (PrimerID, forwardPrimer, Reverse Primer).
Requiement
Preparation
# lower letter to UPPER letter
seqkit seq -u reference.fa > genome.fa
# prepare sequence database for re-PCR searches
famap -t N -b genome.famap genome.fa
fahash -b genome.hash -w 12 -f 3 genome.famapperl ePCR.pl -h############################################################
#
# Usage: ePCR.pl --input primer.txt --output output.txt
#
# Required:
#
# --input <string> input filename, one pair primer per line, tab seperated, e.g.:
# primerID Left_primer_seq Right_primer_seq
#
# --output <string> output filename.
#
############################################################A sliding window function in R. The R package tidyverse should be installed. values is a vector containing column names which need be calculted.
source("./slidingWindow.R")
# An example
sldWid <- slidingWindow(df = df, winSize = 1000000, winStep = 200000, groups = "CHROM", position = "POS", values = c("R.R3.depth", "R.qY.depth"), fun = "mean")A R function for calculating accumulation value for a column of a table. For example, a data.frame contain two columns, "chromosome" and "position", this function will calculate the accumulation position of different chromosome, then a list containing a table with a new column "position_add_Up", a vector containing breaks position, a vector containing labels, a vector containing gaps position, will be returned.
source("./addUp.R")
# An example
addUp(df = df, len = len, group = "chromosome", pos = "position", band = 0.01)
addUp(df = df, len = len, group = "chromosome", pos = c("start", "end"), band = 0.01)A R script for differential expression analysis using DESeq2 (with biological replication). You need to prepare three files:
read count matrix file.samples file, tab-delimited text file indicating biological replicate relationships. e.g.
cond_A cond_A_rep1
cond_A cond_A_rep2
cond_B cond_B_rep1
cond_B cond_B_rep2contrasts file, tab-delimited text file containing the pairs of sample comparisons to perform. e.g.
cond_A cond_B
cond_Y cond_ZUsage:
Rscript run_DESeq2.R -husage: run_DESeq2.R [--] [--help] [--opts OPTS] [--matrix MATRIX]
[--samples_file SAMPLES_FILE] [--min_reps MIN_REPS] [--min_cpm
MIN_CPM] [--contrasts CONTRASTS]
Run differential expression analysis using DESeq2.
flags:
-h, --help show this help message and exit
optional arguments:
-x, --opts RDS file containing argument values
-m, --matrix matrix of raw read counts (not normalized!)
-s, --samples_file tab-delimited text file indicating biological
replicate relationships.
--min_reps At least min count of replicates must have cpm
values > min cpm value. [default: 2]
--min_cpm At least min count of replicates must have cpm
values > min cpm value. [default: 1]
-c, --contrasts file (tab-delimited) containing the pairs of
sample comparisons to perform.If there is a gene/transcript id you are interested and the corresponding genomics data, then you want to abtain genomic, CDS, pep sequence and gene structure information of this gene/transcript, you can use this shll script. Usage:
#chmod u+x extractSeq.sh
./extractSeq.sh --helpUsage: /home/wangpf/bin/extractSeq.sh [--genome genome_file] [--gff3 gff3_file] [--cds cds_file] [--pep pep_file] [--id gene/mRNA_id] [--up up] [--down down] [--gz]
Options:
--genome Specify the genome fasta file
--gff3 Specify the gff3 file
--cds Specify the cds fasta file
--pep Specify the pep fasta file
--id Specify the gene/mRNA id
--up Specify how many bp upstream for gene/mRNA
--down Specify how many bp downstream for gene/mRNA
--gz Compress all the result if this option is present
--help Display this help messageA R script for differential expression analysis using edgeR (without biological replication). You need to prepare three files:
read count matrix file.samples file, tab-delimited text file indicating biological replicate relationships. e.g.
cond_A sample_A
cond_B sample_Bcontrasts file, tab-delimited text file containing the pairs of sample comparisons to perform. e.g.
cond_A cond_B
cond_Y cond_ZUsage:
Rscript run_edgeR.R -husage: run_edgeR.R [--] [--help] [--opts OPTS] [--matrix MATRIX]
[--samples_file SAMPLES_FILE] [--min_reps MIN_REPS] [--min_cpm
MIN_CPM] [--contrasts CONTRASTS] [--dispersion DISPERSION]
Run differential expression analysis using DESeq2.
flags:
-h, --help show this help message and exit
optional arguments:
-x, --opts RDS file containing argument values
-m, --matrix matrix of raw read counts (not normalized!)
-s, --samples_file tab-delimited text file indicating biological
replicate relationships.
--min_reps At least min count of replicates must have cpm
values > min cpm value. [default: 1]
--min_cpm At least min count of replicates must have cpm
values > min cpm value. [default: 1]
-c, --contrasts file (tab-delimited) containing the pairs of
sample comparisons to perform.
-d, --dispersion edgeR dispersion value. [default: 0.1]In some genomics data, there are multiple isoforms fo one gene because of alternative splicing. This perl script can get the longest CDS or pep sequence of genes.
Requiement
- perl module
- Bio::SeqIO
- data file
- CDS or pep sequence file
- gff file
Usage:
perl get_longest_seq.pl -h############################################################
#
# Usage: /public/home/wangpf/bin/get_longest_seq.pl --fasta <cds_or_pep.fa> --gff <genes.gff> --out <outprefix>
#
# Required:
#
# --fasta <string> CDS or pep fasta file.
#
# --gff <string> gff file.
#
# --out <string> output prefix.
#
############################################################
Output
- <outprefix>.longest.fa, the longest CDS or pep sequence in fasta format with gene id as sequence identifier
- <outprefix>.longest.list, a list contain gene id, longest mRNA id and the length of longest mRNA
There are gaps in majority genome fasta files except T2T genome. This find_gaps.py script will find gaps position.
Requiement
- python3 module
- argparse
- Biopython
- input data
- genome fasta file
Usage:
chmod u+x find_gaps.py
find_gaps.py -husage: find_gaps.py [-h] -i INPUT -o OUTPUT [-s] [-c CONTIG]
Find gaps (N regions) in a genome FASTA file and optionally split at gaps.
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
Input genome FASTA file
-o OUTPUT, --output OUTPUT
Output file for gap positions
-s, --split Split the FASTA file at gaps
-c CONTIG, --contig CONTIG
Output file for split contigs (default: split.fa)
Output A tab-separated text file containing three columes.
<chromosome_id> <gap_start> <gap_end>
A fasta file with split contigs.