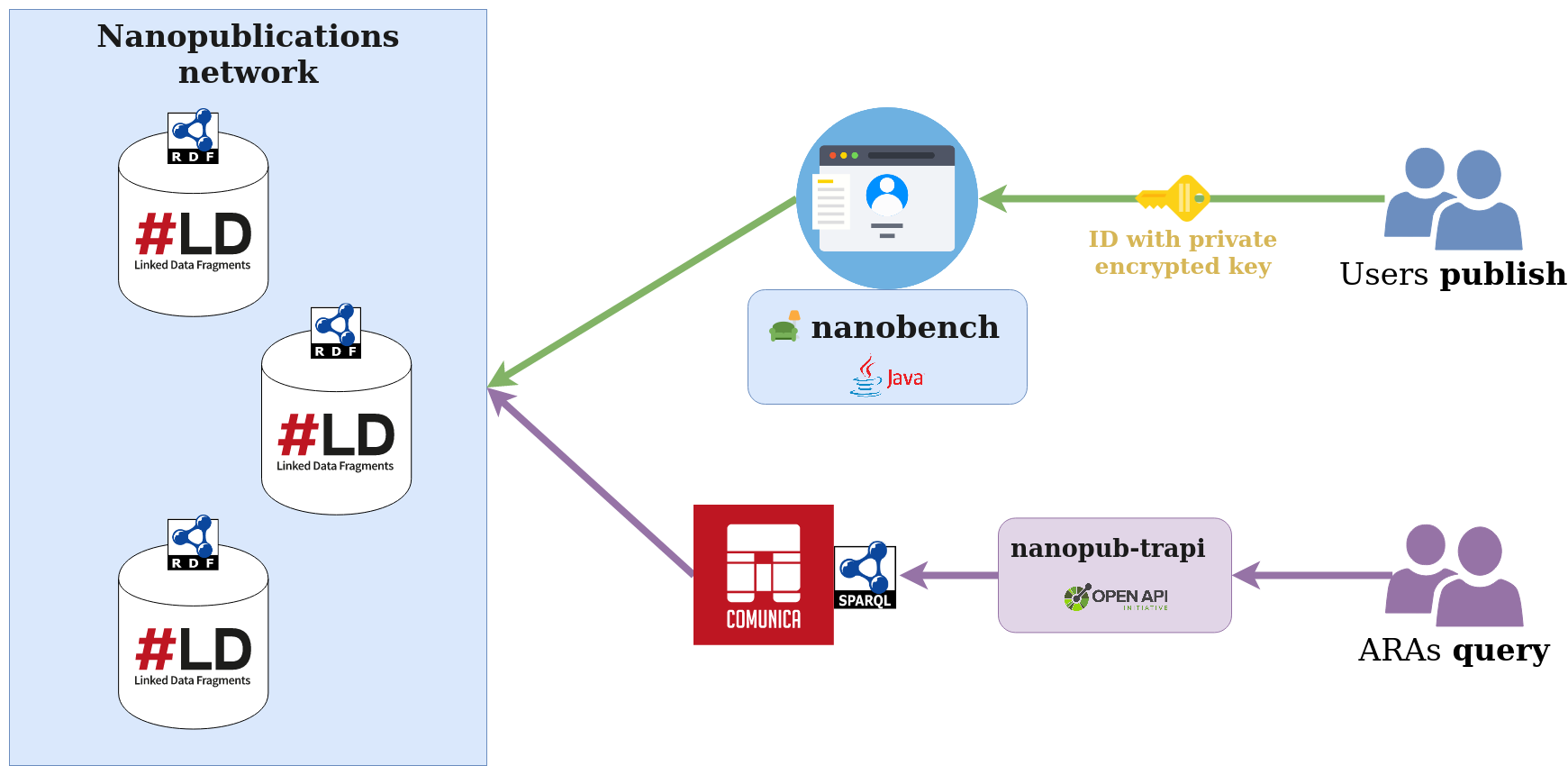
This repository provides guidelines to publish Nanopublications as a user with the Nanobench, and query the published Nanopublications network as a researcher searching for answers 💬
Services has been been deployed publicly to query the Nanopublications network using Translator standards to retrieve the Knowledge Collaboratory graph, a collection of drug indications annotated using preferred identifiers (usually from MONDO, CHEBI, DrugBank, etc).
Check the Translator Reasoner API to query the Nanopublications network SPARQL endpoint at
🔗 https://api.collaboratory.semanticscience.org
- Supports Translator Reasoner API
1.1.0 - Include operation to retrieve the Knowledge Collaboratory nanopublications in the
kgxformat
Requirements: Java 8+
Use the Nanobench 🛋️ to publish and explore nanopublications using a convenient web UI.
Download the latest release of the nanobench.zip file from GitHub: https://github.com/peta-pico/nanobench/releases/tag/nanobench-1.19
Unzip the file, and put the jar file in a convenient folder (at ~/.nanopub/nanobench.jar for example, to be able to call it from anywhere easily)
Visit the complete nanobench installation instructions for more details.
Run the Nanobench on http://localhost:37373. It will use the ids_rsa key in the .nanopub folder to authenticate, or guide you to generate one:
java -jar ~/.nanopub/nanobench.jar -httpPort 37373 -resetExtractSee also:
- Check the vemonet/nanobench wiki to get a full tutorial to publish associations!
- Templates for the Translator (e.g. "Defining a biomedical association with its context") can be seen and improved in the MaastrichtU-IDS/nanobench-templates GitHub repository.
Check the BioLink JSON-LD Context file to find ontologies supported by the Translator (for better integrations with other Translator tools)
Checkout the src/nanopubs/drug_indications_from_gdocs.py file to see an example for publishing drug indications from a gdocs spreadsheet as Nanopublications using the BioLink model
- Clone this repository:
git clone https://github.com/MaastrichtU-IDS/knowledge-collaboratory-api
cd knowledge-collaboratory-api- Install the required dependencies:
pip3 install -r requirements.txt- Test the script to generate nanopubs from the gdoc spreadsheet, and inspect the RDF generated:
python3 src/nanopubs/drug_indications_from_gdocs.pyWhen you are ready, you can run and publish each row of the spreadsheet as a nanopublication:
python3 src/nanopubs/drug_indications_from_gdocs.py --publish --count 10Overview of the different operations available in the Knowledge Collaboratory Translator Reasoner API (supporting kgx)
The user sends a ReasonerAPI query to the Knowledge Collaboratory Nanopublications in the BioLink format (e.g. drug indications). The query is a graph with nodes and edges defined in JSON, and uses classes from the BioLink model.
The /predicates operation will return the entities and relations provided by this API in a JSON object (following the ReasonerAPI specifications).
Experimental, the /kgx operation will return the complete Knowledge Collaboratory drug indications in KGX TSV format (in a .zip file)
-
Drugs indications from the PREDICT publication
-
Drugs indications from the NeuroDKG, and off-label drug indications
Starts the Translator Reasoner API to query the Nanopublications SPARQL endpoint
- Query the Knowledge Collaboratory Nanopublications (drug indications in the BioLink format) using the ReasonerAPI standards and KGX
- Supports Translator Reasoner API
1.1.0 - Include operation to retrieve the Knowledge Collaboratory nanopublications in the
kgxformat - The TRAPI-SPARQL interface and
kgxtransformer are implemented in Python in thesrc/folder
- Supports Translator Reasoner API
- OpenAPI 3 with Swagger UI, built in Python using zalando/connexion
Available at https://api.collaboratory.semanticscience.org 🔗
Starts the Translator Reasoner API to query the Nanopublications SPARQL endpoint, supporting kgx and TRAPI 1.1.0 (defined in this repo in src/)
Requires Python 3.8+ and pip
Clone the repository first
git clone https://github.com/MaastrichtU-IDS/knowledge-collaboratory-api.git
cd knowledge-collaboratory-apiInstall dependencies
pip3 install -e .If you are facing conflict with already installed packages, then you might want to use a Virtual Environment to isolate the installation in the current folder before installing knowledge-collaboratory-api:
# Create the virtual environment folder in your workspace python3.8 -m venv .venv # Activate it using a script in the created folder source .venv/bin/activate
Start the API in production mode on http://localhost:8808:
uvicorn src.main:app --port 8808Or start the API in debug mode (the API will be reloaded automatically at each change to the code):
uvicorn src.main:app --port 8808 --reloadCheck CONTRIBUTING.md for more details on how to run the API locally and contribute.
Requirements: Docker.
Build and start the container with docker-compose 🐳
docker-compose up -d --buildAccess the Swagger UI at http://localhost:8808
We use nginx-proxy and docker-letsencrypt-nginx-proxy-companion as reverse proxy for HTTP and HTTPS in production. You can change the proxy URL and port via environment variables
VIRTUAL_HOST,VIRTUAL_PORTandLETSENCRYPT_HOSTin the docker-compose.yml file.
Check the logs:
docker-compose logsStop the container:
docker-compose downSee the TESTING.md file for more details on testing the API.
Service funded by the NIH NCATS Translator project.